Abstract
Background
The homeobox genes are a large and diverse group of genes, many of which play important roles in the embryonic development of animals. Increasingly, homeobox genes are being compared between genomes in an attempt to understand the evolution of animal development. Despite their importance, the full diversity of human homeobox genes has not previously been described.
Results
We have identified all homeobox genes and pseudogenes in the euchromatic regions of the human genome, finding many unannotated, incorrectly annotated, unnamed, misnamed or misclassified genes and pseudogenes. We describe 300 human homeobox loci, which we divide into 235 probable functional genes and 65 probable pseudogenes. These totals include 3 genes with partial homeoboxes and 13 pseudogenes that lack homeoboxes but are clearly derived from homeobox genes. These figures exclude the repetitive DUX1 to DUX5 homeobox sequences of which we identified 35 probable pseudogenes, with many more expected in heterochromatic regions. Nomenclature is established for approximately 40 formerly unnamed loci, reflecting their evolutionary relationships to other loci in human and other species, and nomenclature revisions are proposed for around 30 other loci. We use a classification that recognizes 11 homeobox gene 'classes' subdivided into 102 homeobox gene 'families'.
Conclusion
We have conducted a comprehensive survey of homeobox genes and pseudogenes in the human genome, described many new loci, and revised the classification and nomenclature of homeobox genes. The classification scheme may be widely applicable to homeobox genes in other animal genomes and will facilitate comparative genomics of this important gene superclass.
Background
Homeobox genes are characterized by the possession of a particular DNA sequence, the homeobox, which encodes a recognizable although very variable protein domain, the homeodomain [1,2]. Most homeodomains are 60 amino acids in length, although exceptions are known. Many homeodomain proteins are transcription factors with important roles in embryonic patterning and cell differentiation, and several have been implicated in human diseases and congenital abnormalities [3].
The homeobox genes have been variously subdivided into superclasses, classes, subclasses or groups, although there has been much inconsistency in the use of these terms. The most commonly recognized groupings are the ANTP, PRD, LIM, POU, HNF, SINE, TALE, CUT, PROS and ZF groups (or variants of these names), although these are not always given equal rank in classification schemes [1,2,4-8]. There is more consensus in classification at a lower level, just above the level of the gene, where very similar genes are grouped into gene families. Widely recognized gene families include Dlx, Evx, Msx, Cdx, En, Otx, Pitx, Otx and Emx (or variants of these names), amongst many others, although there is variation particularly concerning how many gene families are used for the HOX, PAX and NK homeobox genes. Despite the numerous discrepancies, the common principle of classification is the same. The goal of any scheme is to mirror evolutionary diversification, so that 'closely related' genes are placed in the same gene family, and related gene families are placed in the same gene class or other higher grouping. It should be borne in mind, however, that the pathway of evolutionary diversification is never completely known for any large and complex set of genes.
The initial analyses of the draft human genome sequence published in 2001 included estimates of the number of human homeobox genes. Venter et al [9] found 160 homeobox genes, containing 178 homeobox sequences, using large-scale automated classification; while the IHGSC team [10] gave a much higher estimate of 267 homeobox genes. Both were based on draft coverage of the human genome and would be expected to be missing some genes, as well as confusing pseudogenes with genes. In the same year, Banerjee-Basu and Baxevanis [8] presented an analysis of 129 human homeodomain sequences, but this was far from a comprehensive survey. More recently, there have been two more accurate surveys of homeobox genes in the human genome. Nam and Nei [11] found 230 homeobox genes, containing 257 homeobox sequences. Ryan et al [7] found 228 homeodomain sequences in the NCBI RefSeq database of October 2004. Our analyses (described here) revealed many homeobox genes that were incorrectly annotated, named or classified and many homeobox pseudogenes that had previously been missed. We report a complete survey of homeobox loci in the euchromatic regions of the human genome, appropriate gene nomenclature and a consistent classification scheme.
Results and Discussion
How many homeobox genes and pseudogenes?
Using exhaustive database screening, followed by manual examination of sequences, we identified 300 homeobox loci in the human genome. Distinguishing which of these loci are functional genes and which are non-functional pseudogenes was difficult in some cases. Most loci classified as pseudogenes in this study are integrated reverse-transcribed transcripts, readily recognized by their dispersed genomic location, complete lack of intron sequences, and (in some cases) 3' homopolymeric run of adenine residues. A small minority are duplicated copies of genes, recognized by physical linkage to their functional counterparts and the same (or similar) exon-intron arrangement. In general, retrotransposed gene copies are non-functional (and therefore pseudogenes) from the moment of integration because they lack 5' promoter regions necessary for transcription. However, such sequences can occasionally acquire new promoters and become functional as 'retrogenes'. Duplicated gene copies often possess 5'promoter regions (as they are often encompassed by the duplication event); most degenerate to pseudogenes due to redundancy in a process known as non-functionalization, however some can be preserved as functional genes through sub- or neo-functionalization. Thus, in both instances, reliable indicators of non-functionality were sought in order to assign pseudogene status, notably frameshift mutations, premature stop codons and non-synonymous substitutions at otherwise conserved sites in the original coding region.
We currently estimate that the 300 human homeobox loci comprise 235 functional genes and 65 pseudogenes (Table 1). These figures include three functional genes that possess partial homeobox sequences (PAX2, PAX5 and PAX8) and retrotransposed pseudogenes that correspond to only part of the original transcript, whether or not it includes the homeobox region or indeed any of the original coding region. Consequently, 13 retrotransposed pseudogenes that lack homeobox sequences are included (NANOGP11, TPRX1P1, TPRX1P2, POU5F1P7, POU5F1P8, IRX4P1, TGIF2P2, TGIF2P3, TGIF2P4, CUX2P1, CUX2P2, SATB1P1, ZEB2P1). We do not include PAX1, PAX9 and CERS1; these are functional genes without homeobox motifs, albeit closely related to true homeobox genes (the other PAX and CERS genes).
Table 1.
Numbers of human genes, pseudogenes and gene families in each homeobox gene class. The human homeobox gene superclass contains a total of 235 probable functional genes and 65 probable pseudogenes. These are divided between 102 gene families, which are in turn divided between eleven gene classes.
| Class | Subclass | Number of gene families | Number of genes | Number of pseudogenes |
| ANTP | HOXL | 14 | 52 | 0 |
| NKL | 23 | 48 | 19b | |
| PRD | PAX | 3 | 7a | 0 |
| PAXL | 28 | 43 | 24c, d | |
| LIM | 6 | 12 | 0 | |
| POU | 7 | 16 | 8e | |
| HNF | 2 | 3 | 0 | |
| SINE | 3 | 6 | 0 | |
| TALE | 6 | 20 | 10f | |
| CUT | 3 | 7 | 3g | |
| PROS | 1 | 2 | 0 | |
| ZF | 5 | 14 | 1h | |
| CERS | 1 | 5i | 0 | |
| Totals | 102 | 235a | 65b-h | |
aIncludes PAX2, PAX5 and PAX8 that have a partial homeobox; excludes PAX1 and PAX9 that lack a homeobox.
bIncludes NANOGP11 that lacks a homeobox.
cExcludes intronless and repetitive DUX1 to DUX5 sequences.
dIncludes TPRX1P1 and TPRX1P2 that lack a homeobox.
eIncludes POU5F1P7 and POU5F1P8 that lack a homeobox.
fIncludes IRX4P1, TGIF2P2, TGIF2P3 and TGIF2P4 that lack a homeobox.
gIncludes CUX2P1, CUX2P2 and SATB1P1 that lack a homeobox.
hIncludes ZEB2P1 that lacks a homeobox.
iExcludes CERS1 that lacks a homeobox.
The total number of homeobox sequences in the human genome is higher than 300 for two reasons. First, several genes and pseudogenes possess more than one homeobox sequence, notably members of the Dux (double homeobox), Zfhx and Zhx/Homez gene families. Second, we have excluded a set of sequences related to human DUX4 (DUX1 to DUX5), which have become part of 3.3 kb repetitive DNA elements present in multiple copies in the genome [12-14]. Few of these tandemly-repeated sequences are likely to be functional as expressed proteins, and all were probably derived by retrotransposition from functional DUX gene transcripts (see below). The fact that they are not included in the total count, therefore, is likely to have limited bearing on understanding the diversity and normal function of human homeobox genes. Hence, our figure of 300 homeobox loci is the most useful current estimate of the repertoire of human homeobox genes and pseudogenes.
Classification
We propose a simple classification scheme for homeobox genes, based on two principal ranks: gene class and gene family. A gene class contains one or more gene families, which in turn will contain one or more genes. In a few cases, it is useful to erect an intermediate rank between these levels, and for this we use the term subclass. For the entire set of homeobox genes, we use the term superclass.
For the rank of gene family, we use a specific evolutionary-based definition based on common practice in the field of comparative genomics and developmental biology. We define a gene family as a set of genes derived from a single gene in the most recent common ancestor of bilaterian animals (here defined as the latest common ancestor of Drosophila and human). This definition has been made explicitly in previous work [2,6] but is actually a principle that has been in widespread, but rather inconsistent, use for over a decade [15]. For example, amongst the homeobox genes, the En (engrailed) gene family was originally defined to include human EN1 and EN2, plus Drosophila en and inv [16]; these four genes arose by independent duplication from a single gene in the most recent common ancestor of insects and vertebrates. Moving outside the homeobox genes, this principle is also widespread; for example, the Hh (hedgehog) gene family was defined to include mouse Shh, Dhh and Ihh, plus Drosophila hh [17]. To clarify boundaries between gene families, we conducted molecular phylogenetic analyses of human homeodomain sequences, using a range of protostome and occasionally cnidarian homeodomain sequences as outgroups (Additional files 1 and 2).
While the gene family definition described above is generally workable for homeobox genes, by necessity there are some exceptions. One type of exception relates to genes with an unknown ancestral number. For example, there is uncertainty as to whether there were one or two Dlx (distal-less) genes in the most recent common ancestor of bilaterians; however it is common practice to refer to a single Dlx gene family [18]. Thus, we stick with convention for this set of genes. There is similar uncertainty over the ancestral number of Irx (iroquois) genes [19], and again we treat these as a single gene family. The HOX genes are an interesting case as their precise number in the most recent common ancestor of bilaterians is unknown due to lack of phylogenetic resolution between 'central' genes [20]. Here we divide the HOX genes into seven gene families: the 'anterior' Hox1 and Hox2 gene families, the 'group 3' Hox3 gene family, the 'central' Hox4, Hox5 and Hox6-8 gene families, and the 'posterior' Hox9-13 gene family. Another type of exception relates to 'orphan' genes. These are genes that have been found in one species (for example human) but not in other species, or at least not in a wide diversity of Metazoa. Some of these will be ancient genes that have been secondarily lost from the genomes of some species, in which case these comply with our evolutionary definition of a gene family made above. Others, however, will be rapidly evolving genes that originated from another homeobox gene and then diverged to such an extent that their origins are unclear [21]. Whenever origins are unclear, we must define a new gene family to encompass those genes, even though they may not date back to the latest common ancestor of bilaterians. In these cases, the gene family is erected to recognize a set of distinct genes on the basis of DNA and protein sequence, rather than on evolutionary origins.
Using the aforementioned criteria, we recognize 102 homeobox gene families in the human genome (Table 1). We are aware that other homeobox gene families exist in bilaterians but have been lost from humans (for example, Nk7, Ro, Hbn, Repo and Cmp; [7]), and we recognize that some gene family boundaries will alter as new information is obtained. Nonetheless, at the present time the 102 gene families provide a sound framework for the study of human homeobox genes.
It is much more difficult to propose a rigorous evolutionary definition for the rank of gene class. Every attempt to classify genes above the level of gene family involves a degree of arbitrariness. We define gene classes by taking two principal criteria into account. First, gene classes should ideally be monophyletic assemblages of gene families. To identify probable monophyletic groups of gene families, we conducted molecular phylogenetic analyses of homeodomain sequences, and looked for sets of gene families that group together stably, regardless of the precise composition of the dataset used (Figures 1, 2, 3; Additional files 3, 4, 5). Some gene families were difficult to place from sequence data alone, and were found in different gene classes (or subclasses) depending on the precise dataset analyzed or the phylogenetic method employed. This is perhaps not surprising as trees that encompass many homeobox genes can only be built with a short sequence alignment (the homeodomain); under these conditions, phylogenetic trees can only be used as a guide to possible classification, not the absolute truth. In ambiguous cases, we used the chromosomal location of genes to guide possible resolution between alternative hypotheses. Second, some homeobox gene classes can be characterized by the presence of additional protein domains outside of the homeodomain [2]. Recognized protein domains associated with homeodomains include the PRD domain, LIM domain, POU-specific domain, POU-like domain, SIX domain, various MEINOX-related domains, the CUT domain, PROS domain, and various ZF domains [2].
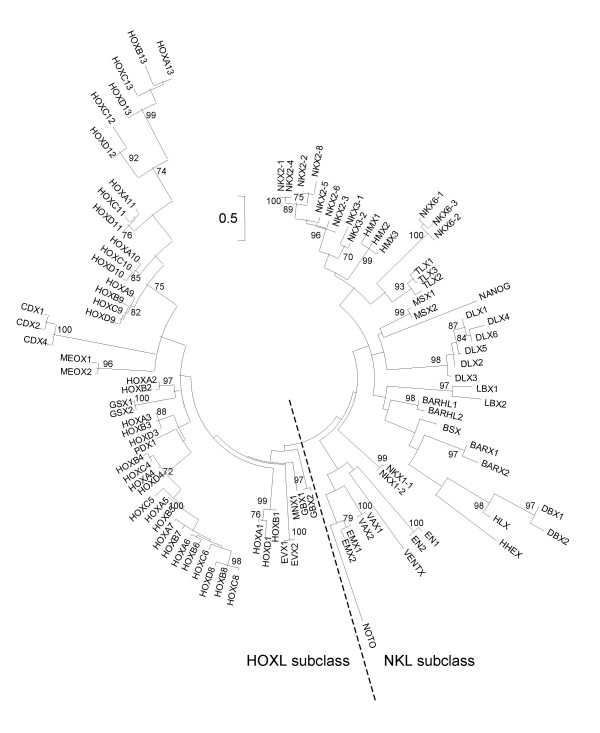
Figure 1.
Maximum likelihood phylogenetic tree of human ANTP-class homeodomains. Arbitrarily rooted phylogenetic tree of human ANTP-class homeodomains constructed using the maximum likelihood method. Bootstrap values supporting internal nodes with over 70% are shown. Homeodomain sequences derived from pseudogenes are excluded. The proposed division between the HOXL and NKL subclasses is indicated. The position of EN1 and EN2 is unstable; this tree places them in the NKL subclass, whereas neighbor-joining analysis of the same dataset places them at the base of the two subclasses (Additional file 3). Interrelationships of genes in the Nk4 and Nk2.2 families are also unstable (in this tree and Additional file 3 respectively); in these cases synteny within and between genomes clearly resolves gene families. Detailed relationships between different gene families should not be inferred from this tree.
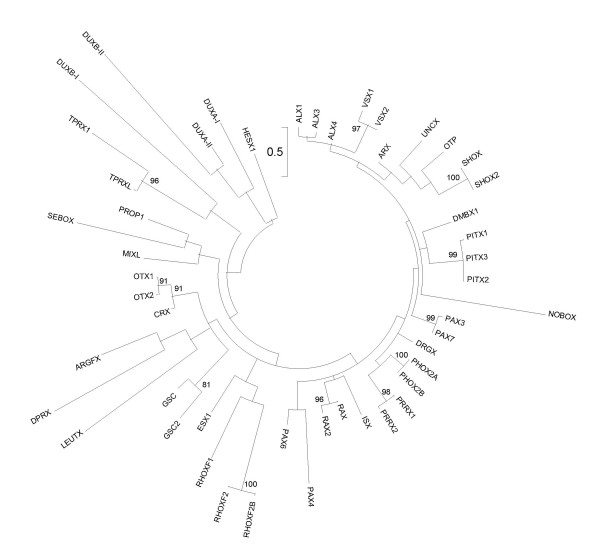
Figure 2.
Maximum likelihood phylogenetic tree of human PRD-class homeodomains. Arbitrarily rooted phylogenetic tree of human PRD-class homeodomains constructed using the maximum likelihood method. Bootstrap values supporting internal nodes with over 70% are shown. Homeodomain sequences derived from pseudogenes are excluded, as are the partial homeodomains of PAX2, PAX5 and PAX8, and the HOPX homeodomain because its extremely divergent sequence destabilizes the overall tree topology. Roman numeral suffixes are used to distinguish multiple homeodomains encoded by a single Dux-family gene. In this tree Dux-family homeodomains are not monophyletic, even within the same gene; however, monophyly is recovered by neighbor-joining analysis (Additional file 4). Detailed relationships between different gene families should not be inferred from this tree.
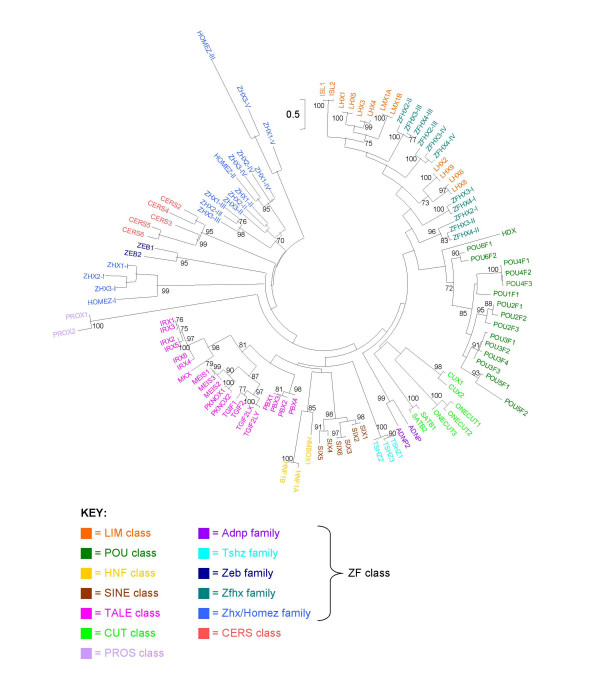
Figure 3.
Maximum likelihood phylogenetic tree of human homeodomains excluding ANTP and PRD classes. Arbitrarily rooted phylogenetic tree of human homeodomains excluding the ANTP and PRD classes constructed using the maximum likelihood method. Bootstrap values supporting internal nodes with over 70% are shown. Homeodomain sequences derived from pseudogenes are excluded. Roman numeral suffixes are used to distinguish multiple homeodomains encoded by a single gene. Classes and/or families are color coded as shown in the key. The LIM and ZF classes are not recovered as two distinct monophyletic groups, a result also found by neighbor-joining analysis (Additional file 5). The multiple homeodomains of Zfhx-family proteins and Zhx/Homez-family proteins are also dispersed in the tree, presumably artefactually. Detailed relationships between different gene families should not be inferred from this tree.
Using the aforementioned criteria, we recognize eleven homeobox gene classes in the human genome: ANTP, PRD, LIM, POU, HNF, SINE, TALE, CUT, PROS, ZF and CERS (Table 1). There is no expectation that the eleven gene classes will be of similar size, simply because some classes will have undergone more expansion by gene duplication than others. In the human genome, the ANTP and PRD classes are much larger than the other classes. Although gene classes should ideally be monophyletic, it is possible that the ZF homeobox gene class, characterized by the presence of zinc finger motifs in most of its members, is polyphyletic (Figure 3; Additional file 5). In other words, domain shuffling may have brought together a homeobox sequence and a zinc finger sequence on more than one occasion. The same may also be true for the LIM class; alternatively the apparent polyphyly of LIM-class homeodomains could be a consequence of LIM domain loss or artefactual placement of some ZF-class homeodomains in phylogenetic analyses (Figure 3; Additional file 5).
In theory, it is possible to recognize higher level associations above the level of the gene class, because the diversification of homeobox genes will have taken place by a continual series of gene duplication events. We do not propose names for hierarchical levels above the rank of class, and consider that gene name, gene family and gene class (and occasionally subclass) convey sufficient information for most purposes.
We use a consistent convention for writing gene classes and gene families. We present the names of all gene classes in abbreviated non-italicized upper case – for example, the ANTP and PRD classes – to avoid confusion with gene symbols (Antp and prd) or indeed gene names (Antennapedia and paired). In contrast, we present the names of all gene families in non-italicized title case; for example, the Cdx, En and Gsc gene families. We have used this style consistently in recent work [6,21-23] and note that several other authors have done likewise [4,7,24]. We suggest that this style, and most of these gene family names, can be used in other bilaterian genomes. Extending the scheme to non-bilaterians is more difficult, however, and awaits clarification of the relationship between the homeobox genes of sponges, placozoans, cnidarians and bilaterians [7,25].
The ANTP homeobox class
The ANTP class derives its name from the Antennapedia (Antp) gene, one of the HOX genes within the ANT-C homeotic complex of Drosophila melanogaster. The human genome has 39 HOX genes, arranged into four Hox clusters. Here we divide the HOX genes into seven gene families: Hox1, Hox2, Hox3, Hox4, Hox5, Hox6-8 and Hox9-13. The HOX genes are not the only ANTP-class genes, and we recognize a total of 37 gene families in this class (Table 1). We divide these 37 gene families between two subclasses that are relatively well-supported in phylogenetic analyses: the HOXL and the NKL subclasses (Figure 1; Additional file 3). As previously discussed, the subclasses are largely consistent with the chromosomal positions of genes [26,27]. The HOXL (HOX-Like or HOX-Linked) genes primarily map to two fourfold paralogous regions: the Hox paralogon (2q, 7p/q, 12q and 17q) and the ParaHox paralogon (4q, 5q, 13q and Xq) (Figure 4). The NKL (NK-Like or NK-Linked) genes are more dispersed, but there is a concentration on the NKL or MetaHox paralogon (2p/8p, 4p, 5q and 10q) (Figure 4). Somewhat aberrantly, the Dlx and En gene families group with the NKL subclass in phylogenetic analyses (Figure 1; Additional file 3), but with the HOXL subclass on the basis of chromosomal positions (Figure 4).
Figure 4.
Chromosomal distribution of human homeobox genes. Ideograms of human chromosomes showing the locations of human homeobox genes. Hox clusters are each shown as a single line for simplicity. Probable pseudogenes are not shown. Genes are color coded according to their class or family (see key). Map positions were obtained through the Ensembl Genome Browser.
Most of the 37 gene families in the ANTP class have been clearly defined before. We draw attention here to several cases that could cause confusion. Other details can be found in Table 2.
Table 2.
Human ANTP class homeobox genes and pseudogenes
| Human ANTP-class homeobox genes and pseudogenes | |||||
| HOXL subclass | |||||
| Family | Gene symbol | Gene name | Location | Entrez gene ID | Previous symbols |
| Cdx | CDX1 | caudal type homeobox 1 | 5q32 | 1044 | |
| CDX2 | caudal type homeobox 2 | 13q12.2 | 1045 | CDX3 | |
| CDX4 | caudal type homeobox 4 | Xq13.2 | 1046 | ||
| Evx | EVX1 | even-skipped homeobox 1 | 7p15.2 | 2128 | |
| EVX2 | even-skipped homeobox 2 | 2q31.1 | 344191 | ||
| Gbx | GBX1 | gastrulation brain homeobox 1 | 7q36.1 | 2636 | |
| GBX2 | gastrulation brain homeobox 2 | 2q37.2 | 2637 | ||
| Gsx | GSX1 | GS homeobox 1 | 13q12.2 | 219409 | GSH1 |
| GSX2 | GS homeobox 2 | 4q12 | 170825 | GSH2 | |
| Hox1 | HOXA1 | homeobox A1 | 7p15.2 | 3198 | HOX1F |
| HOXB1 | homeobox B1 | 17q21.32 | 3211 | HOX2I | |
| HOXD1 | homeobox D1 | 2q31.1 | 3231 | HOX4G | |
| Hox2 | HOXA2 | homeobox A2 | 7p15.2 | 3199 | HOX1K |
| HOXB2 | homeobox B2 | 17q21.32 | 3212 | HOX2H | |
| Hox3 | HOXA3 | homeobox A3 | 7p15.2 | 3200 | HOX1E |
| HOXB3 | homeobox B3 | 17q21.32 | 3213 | HOX2G | |
| HOXD3 | homeobox D3 | 2q31.1 | 3232 | HOX4A | |
| Hox4 | HOXA4 | homeobox A4 | 7p15.2 | 3201 | HOX1D |
| HOXB4 | homeobox B4 | 17q21.32 | 3214 | HOX2F | |
| HOXC4 | homeobox C4 | 12q13.13 | 3221 | HOX3E | |
| HOXD4 | homeobox D4 | 2q31.1 | 3233 | HOX4B | |
| Hox5 | HOXA5 | homeobox A5 | 7p15.2 | 3202 | HOX1C |
| HOXB5 | homeobox B5 | 17q21.32 | 3215 | HOX2A | |
| HOXC5 | homeobox C5 | 12q13.13 | 3222 | HOX3D | |
| Hox6-8 | HOXA6 | homeobox A6 | 7p15.2 | 3203 | HOX1B |
| HOXB6 | homeobox B6 | 17q21.32 | 3216 | HOX2B | |
| HOXC6 | homeobox C6 | 12q13.13 | 3223 | HOX3C | |
| HOXA7 | homeobox A7 | 7p15.2 | 3204 | HOX1A | |
| HOXB7 | homeobox B7 | 17q21.32 | 3217 | HOX2C | |
| HOXB8 | homeobox B8 | 17q21.32 | 3218 | HOX2D | |
| HOXC8 | homeobox C8 | 12q13.13 | 3224 | HOX3A | |
| HOXD8 | homeobox D8 | 2q31.1 | 3234 | HOX4E | |
| Hox9-13 | HOXA9 | homeobox A9 | 7p15.2 | 3205 | HOX1G |
| HOXB9 | homeobox B9 | 17q21.32 | 3219 | HOX2E | |
| HOXC9 | homeobox C9 | 12q13.13 | 3225 | HOX3B | |
| HOXD9 | homeobox D9 | 2q31.1 | 3235 | HOX4C | |
| HOXA10 | homeobox A10 | 7p15.2 | 3206 | HOX1H | |
| HOXC10 | homeobox C10 | 12q13.13 | 3226 | HOX3I | |
| HOXD10 | homeobox D10 | 2q31.1 | 3236 | HOX4D, HOX4E | |
| HOXA11 | homeobox A11 | 7p15.2 | 3207 | HOX1I | |
| HOXC11 | homeobox C11 | 12q13.13 | 3227 | HOX3H | |
| HOXD11 | homeobox D11 | 2q31.1 | 3237 | HOX4F | |
| HOXC12 | homeobox C12 | 12q13.13 | 3228 | HOX3F | |
| HOXA13 | homeobox A13 | 7p15.2 | 3209 | HOX1J | |
| HOXB13 | homeobox B13 | 17q21.32 | 10481 | ||
| HOXC13 | homeobox C13 | 12q13.13 | 3229 | HOX3G | |
| HOXD13 | homeobox D13 | 2q31.1 | 3239 | HOX4I | |
| Mnx | MNX1 | motor neuron and pancreas homeobox 1 | 7q36.3 | 3110 | HLXB9, HB9, HOXHB9 |
| Meox | MEOX1 | mesenchyme homeobox 1 | 17q21.31 | 4222 | MOX1 |
| MEOX2 | mesenchyme homeobox 2 | 7p21.1 | 4223 | MOX2, GAX | |
| Pdx | PDX1 | pancreatic and duodenal homeobox 1 | 13q12.2 | 3651 | IPF1, IUF1, IDX1, STF1 |
| NKL subclass | |||||
| Barhl | BARHL1 | BarH-like homeobox 1 | 9q34.13 | 56751 | |
| BARHL2 | BarH-like homeobox 2 | 1p22.2 | 343472 | ||
| Barx | BARX1 | BARX homeobox 1 | 9q22.32 | 56033 | |
| BARX2 | BARX homeobox 2 | 11q24.3 | 8538 | ||
| Bsx | BSX | brain specific homeobox | 11q24.1 | 390259 | |
| Dbx | DBX1 | developing brain homeobox 1 | 11p15.1 | 120237 | |
| DBX2 | developing brain homeobox 2 | 12q12 | 440097 | ||
| Dlx | DLX1 | distal-less homeobox 1 | 2q31.1 | 1745 | |
| DLX2 | distal-less homeobox 2 | 2q31.1 | 1746 | TES1 | |
| DLX3 | distal-less homeobox 3 | 17q21.33 | 1747 | ||
| DLX4 | distal-less homeobox 4 | 17q21.33 | 1748 | DLX7, DLX8, DLX9, BP1 | |
| DLX5 | distal-less homeobox 5 | 7q21.3 | 1749 | ||
| DLX6 | distal-less homeobox 6 | 7q21.3 | 1750 | ||
| Emx | EMX1 | empty spiracles homeobox 1 | 2p13.2 | 2016 | |
| EMX2 | empty spiracles homeobox 2 | 10q26.11 | 2018 | ||
| En | EN1 | engrailed homeobox 1 | 2q14.2 | 2019 | |
| EN2 | engrailed homeobox 2 | 7q36.3 | 2020 | ||
| Hhex | HHEX | hematopoietically expressed homeobox | 10q23.33 | 3087 | HEX, PRH, PRHX |
| Hlx | HLX | H2.0-like homeobox | 1q41 | 3142 | HLX1, HB24 |
| Lbx | LBX1 | ladybird homeobox 1 | 10q24.32 | 10660 | LBX1H, HPX6 |
| LBX2 | ladybird homeobox 2 | 2p13.1 | 85474 | ||
| Msx | MSX1 | msh homeobox 1 | 4p16.2 | 4487 | HOX7 |
| MSX2 | msh homeobox 2 | 5q35.2 | 4488 | HOX8, MSH | |
| MSX2P1 | msh homeobox 2 pseudogene | 17q23.2 | 55545 | HPX5, MSX2P | |
| Nanog | NANOG | Nanog homeobox | 12p13.31 | 79923 | |
| NANOGP1 | Nanog homeobox pseudogene 1 | 12p13.31 | 404635 | NANOG2 | |
| NANOGP2 | Nanog homeobox pseudogene 2 | 2q36.1 | 414131 | NANOGP4 | |
| NANOGP3 | Nanog homeobox pseudogene 3 | 6p12.1 | 340217 | ||
| NANOGP4 | Nanog homeobox pseudogene 4 | 7p15.1 | 414132 | NANOGP2 | |
| NANOGP5 | Nanog homeobox pseudogene 5 | 9q31.1 | 414133 | ||
| NANOGP6 | Nanog homeobox pseudogene 6 | 10q24.2 | 414134 | ||
| NANOGP7 | Nanog homeobox pseudogene 7 | 14q32.12 | 414130 | NANOGP3 | |
| NANOGP8 | Nanog homeobox pseudogene 8 | 15q14 | 388112 | NANOGP1 | |
| NANOGP9 | Nanog homeobox pseudogene 9 | Xq12 | 349386 | NANOGP6 | |
| NANOGP10 | Nanog homeobox pseudogene 10 | Xp11.3 | 349372 | NANOGP5 | |
| NANOGP11 | Nanog homeobox pseudogene 11 | 6q25.2 | 414135 | ||
| Nk1 | NKX1-1 | NK1 homeobox 1 | 4p16.3 | 54279 | NKX1.1, HSPX153, HPX153 |
| NKX1-2 | NK1 homeobox 2 | 10q26.13 | 390010 | NKX1.2, C10orf121 | |
| Nk2.1 | NKX2-1 | NK2 homeobox 1 | 14q13.3 | 7080 | NKX2.1, NKX2A, TTF1, TITF1 |
| NKX2-4 | NK2 homeobox 4 | 20p11.22 | 4823 | NKX2.4, NKX2D | |
| Nk2.2 | NKX2-2 | NK2 homeobox 2 | 20p11.22 | 4821 | NKX2.2, NKX2B |
| NKX2-8 | NK2 homeobox 8 | 14q13.3 | 26257 | NKX2.8, NKX2H | |
| Nk3 | NKX3-1 | NK3 homeobox 1 | 8p21.2 | 4824 | NKX3.1, NKX3A |
| NKX3-2 | NK3 homeobox 2 | 4p15.33 | 579 | NKX3.2, NKX3B, BAPX1 | |
| Nk4 | NKX2-3 | NK2 homeobox 3 | 10q24.2 | 159296 | NKX2.3, NKX2C, NKX4-3, CSX3 |
| NKX2-5 | NK2 homeobox 5 | 5q35.1 | 1482 | NKX2.5, NKX2E, NKX4-1, CSX, CSX1 | |
| NKX2-6 | NK2 homeobox 6 | 8p21.2 | 137814 | NKX2.6, NKX4-2, CSX2 | |
| Nk5/Hmx | HMX1 | H6 family homeobox 1 | 4p16.1 | 3166 | NKX5-3, H6 |
| HMX2 | H6 family homeobox 2 | 10q26.13 | 3167 | NKX5-2, H6L | |
| HMX3 | H6 family homeobox 3 | 10q26.13 | 340784 | NKX5-1 | |
| Nk6 | NKX6-1 | NK6 homeobox 1 | 4q21.23 | 4825 | NKX6.1, NKX6A |
| NKX6-2 | NK6 homeobox 2 | 10q26.3 | 84504 | NKX6.2, NKX6B, GTX | |
| NKX6-3 | NK6 homeobox 3 | 8p11.21 | 157848 | NKX6.3 | |
| Noto | NOTO | notochord homeobox | 2p13.2 | 344022 | |
| Tlx | TLX1 | T-cell leukemia homeobox 1 | 10q24.32 | 3195 | HOX11, TCL3 |
| TLX2 | T-cell leukemia homeobox 2 | 2p13.1 | 3196 | HOX11L1, NCX | |
| TLX3 | T-cell leukemia homeobox 3 | 5q35.1 | 30012 | HOX11L2, RNX | |
| Vax | VAX1 | ventral anterior homeobox 1 | 10q26.11 | 11023 | |
| VAX2 | ventral anterior homeobox 2 | 2p13.3 | 25806 | ||
| Ventx | VENTX | VENT homeobox | 10q26.3 | 27287 | VENTX2, HPX42B |
| VENTXP1 | VENT homeobox pseudogene 1 | Xp21.3 | 139538 | VENTX2P1, NA88A | |
| VENTXP2 | VENT homeobox pseudogene 2 | 13q31.1 | 347975 | VENTX2P2 | |
| VENTXP3 | VENT homeobox pseudogene 3 | 12q21.1 | 349814 | VENTX2P3 | |
| VENTXP4 | VENT homeobox pseudogene 4 | 3p24.2 | 152101 | VENTX2P4 | |
| VENTXP5 | VENT homeobox pseudogene 5 | 8p12 | 442384 | ||
| VENTXP6 | VENT homeobox pseudogene 6 | 8q21.11 | 552879 | ||
| VENTXP7 | VENT homeobox pseudogene 7 | 3p24.3 | 391518 | VENTX1, HPX42 | |
Human ANTP class homeobox genes and pseudogenes including full names, chromosomal locations, Entrez Gene IDs and previous symbols. NANOGP1 is a duplicate of NANOG.
∘ Cdx, Gsx and Pdx gene families. Some authors refer to the Pdx gene family as the Xlox gene family [28]. One gene from each of these families (CDX2, GSX1 and PDX1) forms the ParaHox cluster at 13q12.2 (Figure 4), and clustering of Cdx, Gsx and Pdx genes is ancestral for chordates [28].
∘ Mnx gene family. This gene family name derives from a previous study [29]. The family includes one gene in the human genome: MNX1 (formerly HLXB9), and two genes in the chicken genome: Mnx1 (formerly HB9) and Mnx2 (formerly MNR2). Some authors refer to the Mnx gene family as the Exex gene family due to the Drosophila ortholog exex [7].
∘ Dlx gene family. It is currently unclear if this gene family is derived from one or more genes in the common ancestor of bilaterians [18]. Phylogenetic analyses place this gene family firmly within the NKL subclass (Figure 1; Additional file 3), but chromosomal positions (on the Hox chromosomes 2, 7 and 17) place it within the HOXL subclass (Figure 4). Here we favor placement of the Dlx gene family within the NKL subclass due to strong phylogenetic support.
∘ En gene family. Phylogenetic analyses place this gene family either within the NKL subclass (maximum likelihood; Figure 1) or close to the division between the NKL and HOXL subclasses (neighbor-joining; Additional file 3). Here we place the En gene family within the NKL subclass, although we note that human EN2 maps close to the clear HOXL-subclass genes GBX1 and MNX1 on chromosome 7 (Figure 4).
∘ Nk2.1 and Nk2.2 gene families. The genes NKX2-1 (formerly TITF1), NKX2-4, NKX2-2 and NKX2-8 divide into two distinct gene families each with an invertebrate ortholog, not a single Nk2 gene family. NKX2-1 and NKX2-4 are collectively orthologous to Drosophila scro and amphioxus AmphiNk2-1 [30,31]; these comprise one gene family: Nk2.1. NKX2-2 and NKX2-8 are collectively orthologous to Drosophila vnd and amphioxus AmphiNk2-2 [31,32]; these comprise a second gene family: Nk2.2.
∘ Nk4 gene family. The genes NKX2-3, NKX2-5 and NKX2-6 form a gene family, quite distinct from other human genes that confusingly share the prefix NKX2. These three genes are actually orthologs of Drosophila tin (formerly NK4); they are not orthologs of Drosophila vnd (formerly NK2) or scro [33]. Therefore, they do not belong to the Nk2.1 or Nk2.2 gene families, but belong to a separate Nk4 gene family. As the three gene names have very extensive current usage, it may be difficult for revised names to be used consistently. In this situation, we don't alter the current names, but raise for discussion the possibility of these genes being renamed to the more logical NKX4-1 (NKX2-5), NKX4-2 (NKX2-6) and NKX4-3 (NKX2-3), or to CSX1 (NKX2-5), CSX2 (NKX2-6) and CSX3 (NKX2-3), based on the alternative name CSX1 for NKX2-5 [34].
∘ Noto gene family. This gene family falls close to the division between the ANTP and PRD classes in phylogenetic analyses (Additional files 1 and 2). We favor placement within the ANTP class as the human NOTO gene is chromosomally linked to the clear ANTP-class (NKL-subclass) genes EMX1, LBX2, TLX2 and VAX2 on chromosome 2 (Figure 4), suggesting ancestry by ancient tandem duplication.
Most of the 100 genes in the ANTP class have been adequately named previously. However, several genes were unnamed or misnamed prior to this study. We have updated these as follows.
∘ GSX2 [Entrez Gene ID: 170825] is the second of two human members of the Gsx gene family. This previously unnamed gene has clear orthology to mouse Gsh2, inferred from sequence identity and synteny. We designate the gene GSX2 and revise the nomenclature of the other human member of the family from GSH1 to GSX1 [Entrez Gene ID: 219409], in accordance with homeobox gene nomenclature convention.
∘ MNX1 [Entrez Gene ID: 3110] is the only member of the Mnx gene family in the human genome. This gene was previously known as HLXB9; we rename it MNX1 because it is not part of a series of at least nine related genes.
∘ PDX1 [Entrez Gene ID: 3651] is the only member of the Pdx gene family in the human genome. This gene was previously known as IPF1; we rename it PDX1 because the majority of published studies use this as the gene symbol.
∘ BSX [Entrez Gene ID: 390259] is the only member of the Bsx gene family in the human genome. We designate this previously unnamed gene BSX on the basis of clear orthology to the mouse Bsx gene, inferred from sequence identity and synteny.
∘ DBX1 [Entrez Gene ID: 120237] and DBX2 [Entrez Gene ID: 440097] are the only two members of the Dbx gene family in the human genome. We designate these previously unnamed genes DBX1 and DBX2 on the basis of clear orthology to mouse Dbx1 and Dbx2, inferred from sequence identity and synteny.
∘ NKX1-1 [Entrez Gene ID: 54729] and NKX1-2 [Entrez Gene ID: 390010] are the only two members of the Nk1 gene family in the human genome. These genes were previously known as HSPX153 and C10orf121 respectively; we rename them NKX1-1 and NKX1-2 on the basis of clear orthology to mouse Nkx1-1 and Nkx1-2, inferred from sequence identity and synteny.
∘ NKX2-1 [Entrez Gene ID: 7080] is the first of two human members of the Nk2.1 gene family. This gene was previously known as TITF1; we rename it NKX2-1 to show that it is a member of the Nk2.1 gene family.
∘ NKX2-6 [Entrez Gene ID: 137814] is the third of three human members of the Nk4 gene family. We designate this previously unnamed gene NKX2-6 on the basis of clear orthology to mouse Nkx2-6, inferred from sequence identity and synteny, although nomenclature revision for the entire Nk4 gene family should be discussed (see above).
∘ NKX3-2 [Entrez Gene ID: 579] is the second of two human members of the Nk3 gene family. This gene was previously known as BAPX1; we rename it NKX3-2 to show that it is a member of the Nk3 gene family.
∘ NKX6-3 [Entrez Gene ID: 157848] is the third of three human members of the Nk6 gene family. We designate this previously unnamed gene NKX6-3 on the basis of clear orthology to mouse Nkx6-3, inferred from sequence identity and synteny.
∘ VENTX [Entrez Gene ID: 27287] is the only functional member of the Ventx gene family in the human genome. This gene was previously known as VENTX2. We remove the numerical suffix from this gene symbol because we discovered that the sequence formerly known as VENTX1 is actually a retrotransposed pseudogene derived from this gene. Accordingly, we also replace the VENTX1 symbol with VENTXP7 (see below).
In contrast to the previous descriptions of probable functional genes, there has been much less research on pseudogenes within the ANTP class. Eleven pseudogenes derived from the human NANOG gene have been described previously [22], while four pseudogenes in the Ventx gene family have been reported following routine annotation of the human genome. We have identified two additional Ventx-family pseudogenes (VENTXP5 and VENTXP6), and also found two cases of pseudogenes that were originally mistaken for functional genes (MSX2P1 and VENTXP7). In all cases, we have clarified the origins and organization of these pseudogenes. This research brings the total number of ANTP-class pseudogenes in the human genome to 19.
∘ MSX2P1 [Entrez Gene ID: 55545]. A short cDNA sequence [EMBL: X74862] related to the Msx gene family was reported previously [35]; the former Entrez Gene record labeled HSHPX5 was based on this sequence. This locus was later provisionally called MSX4, as it was distinct from human MSX1 and MSX2, and by synteny it was clearly not the ortholog of mouse Msx3 [27]. It is now clear that this locus was formed by retrotransposition of mRNA from MSX2 and hence we name it MSX2P1. The genomic sequence of MSX2P1 can now be accessed via the Reference Sequence collection [RefSeq: NR_002307]. The pseudogene shares 91% sequence identity with MSX2 mRNA, lacks intronic sequence, and has remnants of a 3' poly(A) tail. It is intriguing, but probably coincidental, that the MSX2P1 pseudogene has integrated at 17q23.2, close to several ANTP-class genes (HOXB cluster, MEOX1, DLX3 and DLX4).
∘ NANOGP1 [Entrez Gene ID: 404635]. We follow Booth and Holland [22] and classify NANOGP1 as a pseudogene that arose by tandem duplication of NANOG. The alternative view, argued by Hart et al [36], is that this locus is a functional gene, and should be named NANOG2. There is evidence for transcription of this locus in human embryonic stem cells [36], and for selection-driven conservation of the open reading frame [37], but as yet no clear evidence for function.
∘ NANOGP8 [Entrez Gene ID: 388112]. We follow Booth and Holland [22] and classify NANOGP8 as a retrotransposed pseudogene. The alternative view, argued by Zhang et al [38], is that this locus is a functional retrogene. There is evidence for transcription and translation of this locus in cancer cell lines and tumors [38], but no evidence yet for a role in normal tissues.
∘ VENTXP1 [Entrez Gene ID: 139538], VENTXP2 [Entrez Gene ID: 347975], VENTXP3 [Entrez Gene ID: 349814] and VENTXP4 [Entrez Gene ID: 152101]. These four VENTX retrotransposed pseudogenes have been reported previously, and were originally known as VENTX2P1 to VENTX2P4. The correction of the VENTX2 gene symbol to simply VENTX (see above) means that each of the pseudogene names should also change; we rename them VENTXP1 to VENTXP4. VENTXP1 is transcribed but due to mutations it can no longer encode a homeodomain protein; it can however encode an antigenic peptide (NA88A) responsible for T-cell stimulation in response to melanoma [39].
∘ VENTXP5 [Entrez Gene ID: 442384]. We designate this previously unnamed sequence VENTXP5 because it is clearly a retrotransposed pseudogene of VENTX. The genomic sequence of VENTXP5 can now be accessed via the Reference Sequence collection [RefSeq: NG_005091]. The pseudogene shares 83% identity with VENTX mRNA (after masking of an Alu element in the parental mRNA sequence), lacks intronic sequence, and has remnants of a 3' poly(A) tail.
∘ VENTXP6 [Entrez Gene ID: 552879]. We designate this previously unannotated sequence VENTXP6 because it is clearly a retrotransposed pseudogene of VENTX. Its lack of annotation may reflect the fact that it is located within an intron of an unrelated and well characterized gene, STAU2. The genomic sequence of VENTXP6 can now be accessed via the Reference Sequence collection [RefSeq: NG_005090]. The pseudogene shares 87% identity with VENTX mRNA (after masking of an Alu element in the parental mRNA sequence) and lacks intronic sequence.
∘ VENTXP7 [Entrez Gene ID: 391518]. A short cDNA sequence [EMBL: X74864] was reported previously and named HPX42 [35]. This was later renamed the VENTX1 gene, after it was found to be related to Xenopus Ventx-family genes. Our analysis of the genomic sequence at this locus reveals that it is actually a retrotransposed pseudogene of the VENTX gene (formerly VENTX2); thus we designate it VENTXP7. The genomic sequence of VENTXP7 can now be accessed via the Reference Sequence collection [RefSeq: NR_002311]. The pseudogene shares 86% identity with VENTX mRNA (after masking of an Alu element in the parental mRNA sequence), lacks intronic sequence, and has remnants of a 3' poly(A) tail.
One other gene could conceivably be included in the ANTP class, but is excluded from our survey. This gene [Entrez Gene ID: 360030; GenBank: AY151139], has been annotated as a homeobox gene and is located just 20 kb from NANOG. However, no homeodomain was detected when the deduced protein was analyzed for conserved domains. Also, secondary structure prediction did not predict the expected organisation of alpha helices. Alignment with the NANOG homeodomain reveals identity of the KQ and WF motifs, either side of the same intron position (44/45), but few other shared residues. It is possible, but unproven, that the locus arose by tandem duplication of part, or all, of the NANOG homeobox gene. This gene has generated two retrotransposed pseudogenes: one at 2q11.2 and another at 12q24.33.
The PRD homeobox class
The PRD class derives its name from the paired (prd) gene of Drosophila melanogaster. In previous studies, the PRD class has been subdivided in several different ways, often based on identify of the amino acid at residue 50 in the homeodomain, for example S50, K50 and Q50. These categories are not monophyletic groupings of genes and so can be misleading if we aim for a classification scheme that reflects evolution [5]. Here we divide the PRD class into two subclasses of unequal size: the PAX subclass (containing seven PAX genes, excluding PAX1 and PAX9), and the PAXL subclass (containing 43 non-PAX genes and many pseudogenes) (Table 1). PAX genes are defined by possession of a conserved paired-box motif, distinct from the homeobox, coding for the 128-amino-acid PRD domain. Of the nine human genes possessing a paired-box (PAX1 to PAX9), only four also contain a complete homeobox (PAX3, PAX7, PAX4 and PAX6). Three genes have a partial homeobox (PAX2, PAX5 and PAX8), while two lack a homeobox entirely (PAX1 and PAX9). Phylogenetic analyses using PAX genes from a range of species suggest that these are secondary conditions, and that the ancestral PAX gene probably possessed both motifs [40]. The PAX genes do not constitute a single gene family, because it is clear that the latest common ancestor of the Bilateria contained four PAX genes. Three of these are ancestors of the PRD-class homeobox gene families Pax2/5/8, Pax3/7 and Pax4/6; the fourth is the ancestor of PAX1 and PAX9. Thus the PAX subclass contains three gene families. We divide the PAXL subclass into 28 gene families, although as explained below not all of these date to the base of the Bilateria. Thus, we recognize a total of 31 gene families in the PRD class (Table 1).
Many of the 31 gene families in the PRD class have been clearly defined before. We draw attention here to newly defined gene families and cases that could cause confusion. Other details can be found in Table 3.
Table 3.
Human PRD class homeobox genes and pseudogenes
| Human PRD-class homeobox genes and pseudogenes | |||||
| Family | Gene symbol | Gene name | Location | Entrez gene IDc | Previous symbols |
| Alx | ALX1 | ALX homeobox 1 | 12q21.31 | 8092 | CART1 |
| ALX3 | ALX homeobox 3 | 1p13.3 | 257 | ||
| ALX4 | ALX homeobox 4 | 11p11.2 | 6059 | ||
| Argfx | ARGFX | arginine-fifty homeobox | 3q13.33 | 503582 | |
| ARGFXP1 | arginine-fifty homeobox pseudogene 1 | 5q23.2 | 503583 | ||
| ARGFXP2 | arginine-fifty homeobox pseudogene 2 | 17q11.2 | 503640 | ||
| Arx | ARX | aristaless related homeobox | Xp21.3 | 170302 | ISSX |
| Dmbx | DMBX1 | diencephalon/mesencephalon brain homeobox 1 | 1p34.1 | 127343 | MBX, OTX3, PAXB |
| Dprx | DPRX | divergent paired-related homeobox | 19q13.42 | 503834 | |
| DPRXP1 | divergent paired-related homeobox pseudogene 1 | 2q32.1 | 503641 | ||
| DPRXP2 | divergent paired-related homeobox pseudogene 2 | 6p21.31 | 503643 | ||
| DPRXP3 | divergent paired-related homeobox pseudogene 3 | 14q13.2 | 503644 | ||
| DPRXP4 | divergent paired-related homeobox pseudogene 4 | 17q11.2 | 503645 | ||
| DPRXP5 | divergent paired-related homeobox pseudogene 5 | 21q22.13 | 503646 | ||
| DPRXP6 | divergent paired-related homeobox pseudogene 6 | Xp11.4 | 503647 | ||
| DPRXP7 | divergent paired-related homeobox pseudogene 7 | Xq23 | 503648 | ||
| Drgx | DRGX | dorsal root ganglia homeobox | 10q11.23 | 644168 | DRG11, PRRXL1 |
| Dux | DUXA | double homeobox A | 19q13.43 | 503835 | |
| DUXAP1 | double homeobox A pseudogene 1 | 2p11.2 | 503630 | ||
| DUXAP2 | double homeobox A pseudogene 2 | 8q22.3 | 503631 | ||
| DUXAP3 | double homeobox A pseudogene 3 | 10q11.21 | 503632 | ||
| DUXAP4 | double homeobox A pseudogene 4 | 10q11.21 | 503633 | ||
| DUXAP5 | double homeobox A pseudogene 5 | 11q23.3 | 503634 | ||
| DUXAP6 | double homeobox A pseudogene 6 | 15q26.1 | 503635 | ||
| DUXAP7 | double homeobox A pseudogene 7 | 20p11.23 | 503636 | ||
| DUXAP8 | double homeobox A pseudogene 8 | 22q11.21 | 503637 | ||
| DUXAP9 | double homeobox A pseudogene 9 | 14qcen | 503638 | ||
| DUXAP10 | double homeobox A pseudogene 10 | 14q11.2 | 503639 | ||
| DUXB | double homeobox B | 16q23.1 | 100033411 | ||
| Esx | ESX1 | ESX homeobox 1 | Xq22.2 | 80712 | ESX1L, ESXR1 |
| Gsc | GSC | goosecoid homeobox | 14q32.13 | 145258 | GSC1 |
| GSC2 | goosecoid homeobox 2 | 22q11.21 | 2928 | GSCL | |
| Hesx | HESX1 | HESX homeobox 1 | 3p14.3 | 8820 | RPX, ANF |
| Hopx | HOPX | HOP homeobox | 4q12 | 84525 | HOP, OB1, LAGY, NECC1, SMAP31 |
| Isx | ISX | intestine specific homeobox | 22q12.3 | 91464 | RAXLX |
| Leutx | LEUTX | Leucine twenty homeobox | 19q13.2 | 342900 | |
| Mix | MIXL | Mix paired-like homeobox | 1q42.12 | 83881 | MIX, MIXL1, MILD1 |
| Nobox | NOBOX | NOBOX oogenesis homeobox | 7q35 | 135935 | OG2, OG2X |
| Otp | OTP | orthopedia homeobox | 5q14.1 | 23440 | |
| Otx | OTX1 | orthodenticle homeobox 1 | 2p15 | 5013 | |
| OTX2 | orthodenticle homeobox 2 | 14q22.3 | 5015 | ||
| OTX2P1 | orthodenticle homeobox 2 pseudogene | 9q21.2 | 100033409 | OTX2P | |
| CRX | cone-rod homeobox | 19q13.32 | 1406 | OTX3 | |
| Pax2/5/8 | PAX2 | paired box 2 | 10q24.31 | 5076 | |
| PAX5 | paired box 5 | 9p13.2 | 5079 | BSAP | |
| PAX8 | paired box 8 | 2q13 | 7849 | ||
| Pax3/7 | PAX3 | paired box 3 | 2q36.1 | 5077 | HUP2 |
| PAX7 | paired box 7 | 1p36.13 | 5081 | HUP1, PAX7B | |
| Pax4/6 | PAX4 | paired box 4 | 7q32.1 | 5078 | |
| PAX6 | paired box 6 | 11p13 | 5080 | ||
| Phox | PHOX2A | paired-like homeobox 2a | 11q13.4 | 401 | PMX2A, ARIX |
| PHOX2B | paired-like homeobox 2b | 4p13 | 8929 | PMX2B, NBPhox | |
| Pitx | PITX1 | pituitary homeobox 1 | 5q31.1 | 5307 | PTX1, POTX, BFT |
| PITX2 | pituitary homeobox 2 | 4q25 | 5308 | PTX2, ARP1, RGS, RIEG, RIEG1 | |
| PITX3 | pituitary homeobox 3 | 10q24.32 | 5309 | PTX3 | |
| Prop | PROP1 | PROP paired-like homeobox 1 | 5q35.3 | 5626 | |
| Prrx | PRRX1 | paired related homeobox 1 | 1q24.3 | 5396 | PRX1, PMX1, PHOX1 |
| PRRX2 | paired related homeobox 2 | 9q34.11 | 51450 | PRX2, PMX2 | |
| Rax | RAX | retina and anterior neural fold homeobox | 18q21.31 | 30062 | RX |
| RAX2 | retina and anterior neural fold homeobox 2 | 19p13.3 | 84839 | QRX, RAXL1 | |
| Rhox | RHOXF1 | Rhox homeobox family, member 1 | Xq24 | 158800 | PEPP1, OTEX |
| RHOXF2 | Rhox homeobox family, member 2 | Xq24 | 84528 | PEPP2 | |
| RHOXF2B | Rhox homeobox family, member 2B | Xq24 | 727940 | PEPP2L | |
| Sebox | SEBOX | SEBOX homeobox | 17q11.2 | 645832 | OG9, OG9X |
| Shox | SHOX | short stature homeobox | Xp22.33/ Yp11.32 | 6473 | SHOXY, GCFX, PHOG |
| SHOX2 | short stature homeobox 2 | 3q25.32 | 6474 | SHOT, OG12, OG12X | |
| Tprx | TPRX1 | tetra-peptide repeat homeobox 1 | 19q13.32 | 284355 | |
| TPRX2P | tetra-peptide repeat homeobox 2 pseudogene | 19q13.32 | 503627 | ||
| TPRX1P1 | tetra-peptide repeat homeobox 1 pseudogene 1 | 10q22.3 | 503628 | ||
| TPRX1P2 | tetra-peptide repeat homeobox 1 pseudogene 2 | 10q22.3 | 503629 | ||
| TPRXL | tetra-peptide repeat homeobox-like | 3p25.1 | 348825 | ||
| Uncx | UNCX | UNC homeobox | 7p22.3 | 340260 | PHD1, UNCX4.1 |
| Vsx | VSX1 | visual system homeobox 1 | 20p11.21 | 30813 | KTCN, RINX |
| VSX2 | visual system homeobox 2 | 14q24.3 | 338917 | RET1, HOX10, CHX10 | |
Human PRD class homeobox genes and pseudogenes including full names, chromosomal locations, Entrez Gene IDs and previous symbols. Pax2/5/8-family genes contain a partial homeobox. RHOXF2B is a duplicate of RHOXF2. TPRX2P is a duplicate of TPRX1.
∘ Argfx, Dprx and Tprx gene families. There are no known invertebrate members of these three gene families. Therefore, these are exceptions to the rule defining gene families as dating to the base of the Bilateria. The Dprx and Tprx gene families may have arisen by duplication and very extensive divergence from CRX, a member of the Otx gene family, during mammalian evolution; origins of ARGFX are obscure [21].
∘ Dux gene family. Members of this gene family are characterized by the presence of two closely-linked homeobox motifs. Most members are intronless sequences present in multiple polymorphic copies within the 3.3 kb family of tandemly repeated elements associated with heterochromatin. These comprise the sequences known as DUX1 to DUX5 reported in previous studies [12-14] and numerous DUX4 copies detected in this study (see below). The absence of introns suggests that these sequences may have originated by retrotransposition from an mRNA transcript, thus they are probably non-functional. There are two noticeable exceptions; these members known as DUXA and DUXB possess introns, thus either one could be the progenitor for the large number of intronless Dux-family sequences found in the human genome. DUXA has spawned 10 retrotransposed pseudogenes and has been described previously [21]. DUXB is described here (see below).
∘ Hopx gene family. Phylogenetic analyses places this gene family, containing a single very divergent homeobox gene HOPX (formerly HOP), either within the PRD class (maximum likelihood; Additional file 1) or close to Zhx/Homez-family genes (neighbor-joining; Additional file 2). We favor placement in the PRD class for three reasons. First, the HOPX homeodomain has highest sequence identity with PRD-class homeodomains (GSC: 38% and PAX6: 36%). Second, the HOPX homeodomain possesses the same combination of residues that are invariably conserved across human PRD-class homeodomains (Additional file 6). Third, the HOPX homeodomain shares the 46/47 intron position seen in many PRD-class homeodomains. HOPX does not map particularly near any other homeobox genes, although the closest is GSX2 in the ANTP class at 4q12 (Figure 4). HOPX is not a typical PRD-class homeobox gene; the homeodomain has a single amino acid insertion between helix I and helix II (Additional file 6), and lacks the ability to bind DNA [41,42].
∘ Leutx gene family. This gene family contains a single gene in the human genome, LEUTX, and no known invertebrate members. We place LEUTX in the PRD class for four reasons. First, there is weak phylogenetic support for this placement (Additional files 1 and 2). Second, the LEUTX homeodomain possesses the same combination of residues that are invariably conserved across human PRD-class homeodomains (except for a leucine at position 20; Additional file 6). Third, the LEUTX homeodomain shares the 46/47 intron position seen in many PRD-class homeodomains. Fourth, the LEUTX gene is located close to the PRD-class genes TPRX1, CRX, DPRX and DUXA on the distal end of the long arm of chromosome 19 (Figure 4). This fourth observation leads us to hypothesize that this gene family arose by tandem duplication and extensive divergence during mammalian evolution.
∘ Nobox gene family. This gene family falls close to the division between the ANTP and PRD classes in both maximum likelihood and neighbor-joining phylogenetic analyses (Additional files 1 and 2). We favor placement within the PRD class because the NOBOX homeodomain has higher sequence identity with PRD-class homeodomains (up to 55%) than with ANTP-class homeodomains (up to 46%). Chromosomal position does not shed light on the issue, as its location at 7q35 is close to both ANTP- and PRD-class genes (Figure 4).
∘ Otx gene family. This very well known gene family was originally considered to contain human OTX1 and OTX2 (and their mouse orthologs) and the Drosophila otd gene [43]. Later, it was shown that the CRX gene is a member of the same gene family, deriving from the same ancestral gene. Thus, CRX could be considered the true OTX3 gene [44]. Unfortunately, the OTX3 symbol was formerly used erroneously for a gene in a different family, now called DMBX1, thus complicating its future use. The gene family name Otx is derived by majority rule from the constituent genes.
∘ Pax2/5/8 gene family. This gene family is also known as Pax group II; it contains PAX2, PAX5 and PAX8, clearly derived from a single ancestral gene [45]. These genes have partial homeoboxes.
∘ Pax3/7 gene family. This gene family is also known as Pax group III; it contains PAX3 and PAX7, clearly derived from a single ancestral gene [46].
∘ Pax4/6 gene family. This gene family is also known as Pax group IV; it contains PAX4 and PAX6. There is confusion as to whether this should be split into two gene families, because invertebrate homologs generally group with PAX6 in phylogenetic analyses and not as an outgroup to the two genes as might be expected. We follow the generally accepted view and group PAX4 and PAX6 into a single gene family, proposing that PAX4 is a divergent member, not an ancient gene [40].
∘ Rhox gene family. The mouse Rhox cluster was first described as comprising twelve X-linked homeobox genes, all selectively expressed in reproductive tissues [47]. Subsequent studies reported a total of 32 genes in the cluster, with the additional genes attributed to recent tandem duplications [48-51]. The human genome contains three homeobox genes at Xq24 that are clearly members of the Rhox gene family based on sequence identity, molecular phylogenetics, intron positions and chromosomal location. These are RHOXF1 (formerly OTEX/PEPP1), RHOXF2 (formerly PEPP2) and RHOXF2B (formerly PEPP2b/PEPP3).
Most of the 50 genes in the PRD class have been adequately named previously. However, several genes were unnamed or misnamed prior to this study. We have updated these as follows.
∘ ALX1 [Entrez Gene ID: 8092] is the first of three human members of the Alx gene family. This gene was previously known as CART1; we rename it ALX1 because it is related to ALX3 and ALX4; all three genes were formed by duplication from a single ancestral invertebrate gene [52].
∘ DRGX [Entrez Gene ID: 117065] is the only member of the newly defined Drgx gene family in the human genome. This gene was previously known as PRRXL1 and DRG11, and there is a clear mouse ortholog (Prrxl1). The symbol PRRXL1 is misleading because it infers membership of the Prrx gene family, containing PRRX1 and PRRX2 in the human genome. Several lines of evidence suggest it belongs to a different gene family. First, this gene (at 10q11.23) is not located in the same paralogon as PRRX1 (1q24.3) and PRRX2 (9q34.11) so they are not three paralogs generated during genome duplication in early vertebrate evolution. Second, it has a completely different exon-intron structure from the Prrx-family genes, and it does not contain a Prrx domain or an OAR domain (present in PRRX1 and PRRX2; [53]). Third, the homeodomain is only 73% identical to PRRX1 and PRRX2 homeodomains, much lower than the 80-100% usually encountered for members of the same gene family in humans. Finally, we have identified the Drosophila ortholog, IP09201. The homeodomains of Drosophila IP09201 and human DRGX form a highly supported monophyletic group in our maximum likelihood (90%; Additional file 1) and neighbor-joining (97%; Additional file 2) phylogenetic analyses. The new symbol DRGX (dorsal root ganglia homeobox) incorporates the root of the former symbol DRG11, referring to expression of the rodent ortholog in dorsal root ganglia neurons [54].
∘ DUXB [Entrez Gene ID: 100033411] is a human member of the Dux (double homeobox) gene family. As previously discussed, most members of this gene family are intronless and are probably derived by retrotransposition of an mRNA transcript from a functional intron-containing Dux gene (or duplication of such an integrant). Booth and Holland [21] described the DUXA gene containing five introns (including one within each homeobox), and noted the existence of a second intron-containing human Dux-family gene provisionally designated DUXB. The DUXB nomenclature is endorsed here. No cDNA or EST sequences have been reported for DUXB.
∘ GSC2 [Entrez Gene ID: 2928] is the second of two human members of the Gsc gene family. This gene was previously known as GSCL; we rename it GSC2 to remove the inadvertent implication that it is not a true gene, and also to reflect the clear orthology to chick Gsc2 as inferred by phylogenetic analysis and synteny.
∘ HOPX [Entrez Gene ID: 84525] is the only member of the newly defined Hopx gene family in the human genome. The mouse version of the gene was first identified first and named Hop (homeodomain only protein) because the encoded protein is just 73 amino acids long, with 61 of these making up the homeodomain [41,42]. The HOP gene symbol is not ideal as it is also used for unrelated genes, including hopscotch in Drosophila and hop-sterile in mouse. Therefore, we revise the gene symbol from HOP to HOPX (HOP homeobox) in accordance with homeobox gene nomenclature convention.
∘ LEUTX [Entrez Gene ID: 342900] is the only member of the newly defined Leutx gene family in the human genome. We designate this previously unnamed gene LEUTX (leucine twenty homeobox) to reflect the presence of a leucine residue at the otherwise highly conserved homeodomain position 20; other PRD-class homeodomains have a phenylalanine at this position (Additional file 6). Studies of mutations in other homeobox genes suggest that mutation to leucine alters transcriptional activity of a homeodomain protein [55].
∘ RAX2 [Entrez Gene ID: 84839] is the second of two human members of the Rax gene family. This gene was previously known as RAXL1; we rename it RAX2 to standardize nomenclature.
∘ RHOXF1 [Entrez Gene ID: 158800] and RHOXF2 [Entrez Gene ID: 84528] are two of three human members of the Rhox gene family. These genes were previously known as OTEX/PEPP1 and PEPP2 respectively. The prefix PEPP is not suitable as it is used for numerous aminopeptidase P-encoding genes. Thus, we replace the gene symbols OTEX/PEPP1 and PEPP2 with RHOXF1 and RHOXF2 respectively, to reflect their orthologous relationship with the mouse Rhox cluster (containing 32 genes, see above) whilst avoiding inadvertent equivalence to specific genes within the cluster.
∘ RHOXF2B [Entrez Gene ID: 727940] is the third human member of the Rhox gene family. This locus was referred to in previous studies as PEPP2b [56] and PEPP3 [51]. The prefix PEPP cannot be approved for reasons noted above. RHOXF2B is located very close to RHOXF1 and RHOXF2 at Xq24 and is clearly a very recent duplicate of RHOXF2. The genomic sequences at these two loci share 99% identity over exonic, intronic and approximately 20 kb flanking regions. Over the coding region, there are just two nucleotide substitutions (both nonsynonymous); one of these results in an unusual change within the homeodomain (arginine to cysteine at position 18). We currently list RHOXF2B as a functional gene, although it is possible that it is a duplicated pseudogene.
∘ SEBOX [Entrez Gene ID: 645832] is the only member of the Sebox gene family in the human genome. The human gene is the ortholog of mouse Sebox based on their locations in syntenic chromosomal regions (17q11.2 and 11B5 respectively) and presence of the same intron positions. However, sequence identity is lower than normal for orthologous genes in mouse and human (78% amino acid identity over the homeodomain) and there is evidence that the human gene has undergone divergence. Most surprisingly, the human sequence has two unusual substitutions in the homeodomain [57]. At homeodomain position 51, the human sequence codes for lysine whereas mouse has asparagine; an earlier analysis of 346 homeodomain sequences found asparagine to be invariant at this position [1,2]. Similarly, at homeodomain position 53, human has tryptophan whereas mouse has arginine; this position is almost invariably arginine [1,2]. These sequence changes in the important third helix raise the possibility that human SEBOX could have accumulated mutations as a non-functional pseudogene. Until this is shown more clearly we consider it to be a functional, but divergent, gene. This gene was previously known as OG9X with SEBOX as the alternative symbol; we favor SEBOX because the OG prefix was originally used for several unrelated homeobox genes.
∘ UNCX [Entrez Gene ID: 340260] is the only member of the Uncx gene family in the human genome. This gene was previously known as UNCX4.1; we remove the numerals to give UNCX as these do not denote a series within a gene family.
∘ VSX2 [Entrez Gene ID: 338917] is the second of two human members of the Vsx gene family. This gene was previously known as CHX10; we rename it VSX2 to better reflect its paralogous relationship to VSX1. VSX2 has been used as an alias for this gene in other vertebrate species and the gene symbol CHX10 has the disadvantage of implicitly suggesting presence of at least nine paralogs in human (CHX1 to CHX9), which do not exist.
Unlike the situation with the ANTP class, many of the pseudogenes within the PRD class have been well characterized. A previous study has described and named two pseudogenes in the Argfx gene family, seven pseudogenes in the Dprx gene family, four pseudogenes in the Tprx gene family, and 10 pseudogenes derived from the DUXA gene [21]. There is also a possibility that the SEBOX and RHOXF2B loci are non-functional pseudogenes, as described above. We have identified a previously undescribed pseudogene from the Otx gene family (OTX2P1), and argue that the majority of Dux-family sequences are pseudogenes.
∘ OTX2P1 [Entrez Gene ID: 100033409]. We designate this previously undescribed sequence OTX2P1 because it is clearly a retrotransposed pseudogene of OTX2. The genomic DNA sequence of OTX2P1 shares significant homology with OTX2 transcript variant 2 [RefSeq: NM_172337]. There is an Alu element (AluSx subfamily) insertion, a Made1 (Mariner derived element 1) insertion, and a 1182-nucleotide deletion in OTX2P1 compared to OTX2. The OTX2P1 sequence lacks introns, ends with a poly(A) tail, and harbors critical sequence alterations (including a three-nucleotide insertion introducing a stop codon into the deduced homeodomain).
∘ DUX1 [EMBL: AJ001481], DUX2 [GenBank: AF068744], DUX3 [GenBank: AF133130] and DUX5 [GenBank: AF133131]. These sequences have been cloned in previous studies [12,13]. We detected no matches with 100% identity to DUX1, DUX2, DUX3 or DUX5 in build 35.1 of the human genome sequence, which covers the euchromatic regions of each chromosome. This concurs with previous studies indicating that DUX1, DUX2, DUX3 and DUX5 are found in heterochromatin on human acrocentric chromosomes; each is apparently present in multiple copies within members of the 3.3 kb family of tandemly repeated DNA elements [12,13]. Because the majority of human heterochromatin has not been sequenced, and may be variable between individuals, the exact number of copies of DUX1, DUX2, DUX3 and DUX5 is unknown. It is also debatable whether these loci encode functional proteins. These sequences lack introns and, as discussed above, are most likely derived from intron-containing genes in the Dux family, such as DUXA or DUXB.
∘ DUX4 [GenBank: AF117653]. This sequence has been extensively studied as some of its multiple copies exist within the 3.3 kb repetitive elements of the D4Z4 locus at 4q35 [14]. The polymorphic D4Z4 locus is linked to facioscapulohumeral muscular dystrophy (FSHD); between 12 and 96 tandem copies of 3.3 kb elements are present in unaffected individuals and deletions leaving a maximum of eight such elements have been associated with FSHD [58]. In build 35.1 of the human genome sequence, we identified 35 loci at 10 chromosomal locations containing a total of 58 DUX4 (and highly similar) homeobox sequences. This should not be taken as a precise figure due to copy number polymorphism and the possibility of additional copies existing in currently unsequenced heterochromatic regions. Some of the copies are 100% identical to the previously reported DUX4 sequence over the homeobox regions, others have single nucleotide polymorphisms, some have critical sequence mutations, and others have just a single homeobox. Most of the copies are located in tandemly repeated arrays (for example, on chromosomes 4, 10 and 16) and others are alone in the genome (for example, a single copy resides at 3p12.3). The majority of DUX4 copies are unlikely to encode functional proteins as suggested by their intronless, mutated and tandemly repeated nature. The lack of introns indicates they are most likely derived from intron-containing genes in the Dux family, such as DUXA or DUXB.
The LIM homeobox class
The LIM class encodes proteins with two LIM domains (named from the nematode lin-11, mammalian Isl1 and nematode mec-3 genes) N-terminal to a typical (i.e. 60-amino-acid) homeodomain. The LIM domain is a protein-protein interaction domain of approximately 55 amino acids comprising two specialised cysteine-rich zinc fingers in tandem [59]. Importantly, human genes also exist that encode LIM domains but not homeodomains. These LIM domains are divergent from the LIM domains encoded by LIM homeobox genes, and hence these genes are unlikely to be derived by loss of the homeobox. There is one exception: the human Lmo gene family encodes LIM domains that have been grouped by sequence similarity and domain arrangement with the LIM domains of the LIM homeobox gene class [59]. Thus, this gene family may have secondarily lost the homeobox, although this remains untested. Only genes encoding both LIM domains and homeodomains are included in our LIM homeobox gene count.
We have identified a total of twelve LIM-class homeobox genes in the human genome (Tables 1 and 4), consistent with previous work [60]. Phylogenetic analyses of homeodomains do not always recover the LIM class as a monophyletic group, depending on the dataset and method used (Figure 3; Additional files 1, 2 and 5), but it is likely that the class evolved from a single fusion event that brought together LIM domains and a homeodomain. Phylogenetic analyses of homeodomains divide the LIM class into six gene families (Figure 3; Additional files 1, 2 and 5), consistent with previous studies [60]. Each gene family has two human members and dates to a single ancestral gene in the most recent common ancestor of bilaterians [60]. We have not found any human LIM-class pseudogenes.
Table 4.
Human LIM, POU, HNF, SINE, TALE, CUT, PROS, ZF AND CERS class homeobox genes and pseudogenes
| Human LIM-class homeobox genes | |||||
| Family | Gene symbol | Gene name | Location | Entrez gene ID | Previous symbols |
| Isl | ISL1 | ISL LIM homeobox 1 | 5q11.2 | 3670 | |
| ISL2 | ISL LIM homeobox 2 | 15q24.3 | 64843 | ||
| Lhx1/5 | LHX1 | LIM homeobox 1 | 17q12 | 3975 | LIM1 |
| LHX5 | LIM homeobox 5 | 12q24.13 | 64211 | ||
| Lhx2/9 | LHX2 | LIM homeobox 2 | 9q33.3 | 9355 | LH2 |
| LHX9 | LIM homeobox 9 | 1q31.3 | 56956 | ||
| Lhx3/4 | LHX3 | LIM homeobox 3 | 9q34.3 | 8022 | M2-LHX3 |
| LHX4 | LIM homeobox 4 | 1q25.3 | 89884 | GSH4 | |
| Lhx6/8 | LHX6 | LIM homeobox 6 | 9q33.2 | 26468 | LHX6.1 |
| LHX8 | LIM homeobox 8 | 1p31.1 | 431707 | LHX7 | |
| Lmx | LMX1A | LMX LIM homeobox 1A | 1q24.1 | 4009 | LMX1, LMX1.1 |
| LMX1B | LMX LIM homeobox 1B | 9q33.3 | 4010 | LMX2, LMX1.2 | |
| Human POU-class homeobox genes and pseudogenes | |||||
| Hdx | HDX | highly divergent homeobox | Xq21.1 | 139324 | CXorf43 |
| Pou1 | POU1F1 | POU class 1 homeobox 1 | 3p11.2 | 5449 | PIT1, GHF1 |
| Pou2 | POU2F1 | POU class 2 homeobox 1 | 1q24.2 | 5451 | OCT1, OTF1 |
| POU2F2 | POU class 2 homeobox 2 | 19q13.2 | 5452 | OCT2, OTF2 | |
| POU2F3 | POU class 2 homeobox 3 | 11q23.3 | 25833 | OCT11, PLA1, EPOC1, SKN1A | |
| Pou3 | POU3F1 | POU class 3 homeobox 1 | 1p34.3 | 5453 | OCT6, OTF6, SCIP |
| POU3F2 | POU class 3 homeobox 2 | 6q16.2 | 5454 | OCT7, OTF7, BRN2, POUF3 | |
| POU3F3 | POU class 3 homeobox 3 | 2q12.1 | 5455 | OTF8, BRN1 | |
| POU3F4 | POU class 3 homeobox 4 | Xq21.1 | 5456 | OTF9, BRN4 | |
| Pou4 | POU4F1 | POU class 4 homeobox 1 | 13q31.1 | 5457 | BRN3A, RDC1, Oct-T1 |
| POU4F2 | POU class 4 homeobox 2 | 4q31.22 | 5458 | BRN3B, BRN3.2 | |
| POU4F3 | POU class 4 homeobox 3 | 5q32 | 5459 | BRN3C | |
| Pou5 | POU5F1 | POU class 5 homeobox 1 | 6p21.33 | 5460 | OCT3, OTF3, OCT4, OTF4 |
| POU5F1P1 | POU class 5 homeobox 1 pseudogene 1 | 8q24.21 | 5462 | OTF3C, OTF3P1, POU5FLC8 | |
| POU5F1P2 | POU class 5 homeobox 1 pseudogene 2 | 8q22.3 | 100009665 | ||
| POU5F1P3 | POU class 5 homeobox 1 pseudogene 3 | 12p13.31 | 642559 | OTF3L, POU5F1L, POU5FLC12 | |
| POU5F1P4 | POU class 5 homeobox 1 pseudogene 4 | 1q22 | 645682 | POU5FLC1 | |
| POU5F1P5 | POU class 5 homeobox 1 pseudogene 5 | 10q21.3 | 100009667 | ||
| POU5F1P6 | POU class 5 homeobox 1 pseudogene 6 | 3q21.3 | 100009668 | ||
| POU5F1P7 | POU class 5 homeobox 1 pseudogene 7 | 3q12.1 | 100009669 | ||
| POU5F1P8 | POU class 5 homeobox 1 pseudogene 8 | 17q25.3 | 100009670 | ||
| POU5F2 | POU class 5 homeobox 2 | 5q15 | 134187 | SPRM1 | |
| Pou6 | POU6F1 | POU class 6 homeobox 1 | 12q13.13 | 5463 | BRN5, MPOU, TCFB1 |
| POU6F2 | POU class 6 homeobox 2 | 7p14.1 | 11281 | WT5, WTSL, RPF1 | |
| Human HNF-class homeobox genes | |||||
| Hmbox | HMBOX1 | homeobox containing 1 | 8p12 | 79618 | HNF1LA, PBHNF |
| Hnf1 | HNF1A | HNF1 homeobox A | 12q24.31 | 6927 | TCF1, HNF1, LFB1 |
| HNF1B | HNF1 homeobox B | 17q12 | 6928 | TCF2, LFB3, VHNF1 | |
| Human SINE-class homeobox genes | |||||
| Six1/2 | SIX1 | SIX homeobox 1 | 14q23.1 | 6495 | |
| SIX2 | SIX homeobox 2 | 2p21 | 10736 | ||
| Six3/6 | SIX3 | SIX homeobox 3 | 2p21 | 6496 | |
| SIX6 | SIX homeobox 6 | 14q23.1 | 4990 | OPTX2, Six9 | |
| Six4/5 | SIX4 | SIX homeobox 4 | 14q23.1 | 51804 | AREC3 |
| SIX5 | SIX homeobox 5 | 19q13.32 | 147912 | DMAHP | |
| Human TALE-class homeobox genes and pseudogenes | |||||
| Irx | IRX1 | iroquois homeobox 1 | 5p15.33 | IRX-5 | |
| IRX1P1 | iroquois homeobox 1 pseudogene 1 | 13q12.12 | 79192 | IRXA1 | |
| IRX2 | iroquois homeobox 2 | 5p15.33 | 646390 | ||
| IRX3 | iroquois homeobox 3 | 16q12.2 | 153572 | IRX-1 | |
| IRX4 | iroquois homeobox 4 | 5p15.33 | 50805 | ||
| IRX4P1 | iroquois homeobox 4 pseudogene 1 | 18p11.22 | 100009671 | ||
| IRX5 | iroquois homeobox 5 | 16q12.2 | 79190 | IRX2A | |
| IRX6 | iroquois homeobox 6 | 16q12.2 | IRX-3, IRX7 | ||
| Meis | MEIS1 | Meis homeobox 1 | 2p14 | 4211 | |
| MEIS2 | Meis homeobox 2 | 15q14 | 4212 | MRG1 | |
| MEIS3 | Meis homeobox 3 | 19q13.32 | 56917 | MRG2 | |
| MEIS3P1 | Meis homeobox 3 pseudogene 1 | 17p12 | 4213 | MRG2, MEIS3, MEIS4 | |
| MEIS3P2 | Meis homeobox 3 pseudogene 2 | 17p11.2 | 257468 | ||
| Mkx | MKX | mohawk homeobox | 10p12.1 | 283078 | IRXL1, IFRX, C10orf48 |
| Pbx | PBX1 | pre-B-cell leukemia homeobox 1 | 1q23.3 | 5087 | |
| PBX2 | pre-B-cell leukemia homeobox 2 | 6p21.32 | 5089 | G17, HOX12, PBX2MHC | |
| PBX2P1 | pre-B-cell leukemia homeobox 2 pseudogene 1 | 3q24 | 5088 | PBXP1, PBX2 | |
| PBX3 | pre-B-cell leukemia homeobox 3 | 9q33.3 | 5090 | ||
| PBX4 | pre-B-cell leukemia homeobox 4 | 19p13.11 | 80714 | ||
| Pknox | PKNOX1 | PBX/knotted homeobox 1 | 21q22.3 | 5316 | PREP1, PKNOX1C |
| PKNOX2 | PBX/knotted homeobox 2 | 11q24.2 | 63876 | PREP2 | |
| Tgif | TGIF1 | TGFB-induced factor homeobox 1 | 18p11.31 | 7050 | TGIF, HPE4 |
| TGIF1P1 | TGFB-induced factor homeobox1 pseudogene 1 | 19q13.32 | 126052 | ||
| TGIF2 | TGFB-induced factor homeobox 2 | 20q11.23 | 60436 | ||
| TGIF2P1 | TGFB-induced factor homeobox 2 pseudogene 1 | 1q44 | 126826 | ||
| TGIF2P2 | TGFB-induced factor homeobox 2 pseudogene 2 | 15q21.1 | 100009674 | ||
| TGIF2P3 | TGFB-induced factor homeobox 2 pseudogene 3 | 15q21.1 | 100009672 | ||
| TGIF2P4 | TGFB-induced factor homeobox 2 pseudogene 4 | 14q24.2 | 100009673 | ||
| TGIF2LX | TGFB-induced factor homeobox 2-like, X-linked | Xq21.31 | 90316 | TGIFLX (retrogene) | |
| TGIF2LY | TGFB-induced factor homeobox 2-like, Y-linked | Yp11.2 | 90655 | TGIFLY (retrogene) | |
| Human CUT-class homeobox genes and pseudogenes | |||||
| Onecut | ONECUT1 | one cut homeobox 1 | 15q21.3 | 3175 | HNF6, HNF6A |
| ONECUT2 | one cut homeobox 2 | 18q21.31 | 9480 | OC2 | |
| ONECUT3 | one cut homeobox 3 | 19p13.3 | 390874 | ||
| Cux | CUX1 | cut-like homeobox 1 | 7q22.1 | 1523 | CUTL1, CUX, CDP, COY1 |
| CUX2 | cut-like homeobox 2 | 12q24.12 | 23316 | CUTL2 | |
| CUX2P1 | cut-like homeobox 2 pseudogene 1 | 10p14 | - | ||
| CUX2P2 | cut-like homeobox 2 pseudogene 2 | 4q32.1 | - | ||
| Satb | SATB1 | SATB homeobox 1 | 3p24.3 | 6304 | |
| SATB2 | SATB homeobox 2 | 2q33.1 | 23314 | ||
| Human PROS-class homeobox genes | |||||
| Prox | PROX1 | prospero homeobox 1 | 1q41 | 5629 | |
| PROX2 | prospero homeobox 2 | 14q24.3 | 283571 | ||
| Human ZF-class homeobox genes and pseudogenes | |||||
| Adnp | ADNP | activity-dependent neuroprotector homeobox | 20q13.13 | 23394 | ADNP1 |
| ADNP2 | ADNP homeobox 2 | 18q23 | 22850 | ZNF508 | |
| Tshz | TSHZ1 | teashirt zinc finger homeobox 1 | 18q22.3 | 10194 | TSH1 |
| TSHZ2 | teashirt zinc finger homeobox 2 | 20q13.2 | 128553 | TSH2, ZNF218, ZABC2, OVC10-2 | |
| TSHZ3 | teashirt zinc finger homeobox 3 | 19q12 | 57616 | TSH3, ZNF537 | |
| Zeb | ZEB1 | zinc finger E-box binding homeobox 1 | 10p11.22 | 6935 | ZFHX1A, deltaEF1, TCF8, ZEB |
| ZEB2 | zinc finger E-box binding homeobox 2 | 2q22.3 | 9839 | ZFHX1B, SIP1, SMADIP1 | |
| ZEB2P1 | zinc finger E-box binding homeobox 2 pseudogene 1 | 4p15.32 | 100033412 | ||
| Zfhx | ZFHX2 | zinc finger homeobox 2 | 14q11.2 | 85446 | |
| ZFHX3 | zinc finger homeobox 3 | 16q22.3 | 463 | ATBT, ATBF1 | |
| ZFHX4 | zinc finger homeobox 4 | 8q21.11 | 79776 | ZFH4 | |
| Zhx/ | ZHX1 | zinc fingers and homeoboxes 1 | 8q24.13 | 11244 | |
| Homez | ZHX2 | zinc fingers and homeoboxes 2 | 8q24.13 | 22882 | |
| ZHX3 | zinc fingers and homeoboxes 3 | 20q12 | 23051 | TIX1 | |
| HOMEZ | homeobox and leucine zipper encoding | 14q11.2 | 57594 | ||
| Human CERS-class homeobox genes | |||||
| Cers | CERS2 | ceramide synthase 2 | 1p36.13-q24.1 | 29956 | LASS2, TRH3, TMSG1 |
| CERS3 | ceramide synthase 3 | 15q26.3 | 204219 | LASS3 | |
| CERS4 | ceramide synthase 4 | 19p13.3 | 79603 | LASS4, TRH1 | |
| CERS5 | ceramide synthase 5 | 12q13.12 | 91012 | LASS5, TRH4 | |
| CERS6 | ceramide synthase 6 | 2q31 | 253782 | LASS6 | |
Human homeobox genes and pseudogenes, excepting the ANTP and PRD classes, including full names, chromosomal locations, Entrez Gene IDs and previous symbols. The HOMEZ gene is in the ZF class but encodes a protein with leucine zippers instead of zinc fingers.
The POU homeobox class
The POU class generally encodes proteins with a POU-specific domain (named from the mammalian genes Pit1 (now Pou1f1), OCT1 and OCT2 (now POU2F1 and POU2F2), andnematode unc-86) N-terminal to a typical homeodomain. The POU-specific domain is a DNA-binding domain of approximately 75 amino acids; the POU-specific domain and the homeodomain are collectively known as the bipartite POU domain [61].
We have identified a total of 16 POU-class homeobox genes in the human genome (Tables 1 and 4). The genes form a distinct grouping even if the POU-specific domain is disregarded – phylogenetic analyses of homeodomains recover the POU class as a monophyletic group (Figure 3; Additional files 1, 2 and 5). There are six widely recognized gene families within the POU class (Pou1 to Pou6), and nomenclature revisions approximately 10 years ago clarified which genes belong to which gene family [62]. We have placed two additional genes (HDX and POU5F2) in the POU class on the basis of their deduced homeodomain sequences, even though one of these genes (HDX) does not encode a POU-specific domain. We have erected a new gene family for this gene, bringing the total number of gene families in the POU class to seven. We have also identified a total of eight POU-class pseudogenes in the human genome (Tables 1 and 4); we have named six of these (POU5F1P2, POU5F1P4 to POU5F1P8), and revised the nomenclature of one other (POU5F1P3).
∘ HDX [Entrez Gene ID: 139324]. This gene was previously known as CXorf43. The gene encodes a highly divergent atypical (68-amino-acid) homeodomain but not a POU-specific domain, and thus it is debatable whether it should be placed within the POU class. Phylogenetic analyses of homeodomains place it basally in a clade with the POU class (Figure 3; Additional files 1 and 5), or within the POU class (Additional file 2), suggesting that the HDX protein either diverged before the POU-specific domain became associated with the homeodomain or lost the POU-specific domain during evolution. Further information on this gene may allow this tentative classification to be revisited.
∘ POU5F2 [Entrez Gene ID: 134187]. We designate this previously unnamed gene POU5F2 on the basis of clear orthology to the mouse Sprm1 gene, which has been assigned the second member of the Pou5 gene family [63]. The symbol POU5F2 ensures the gene conforms with standardized nomenclature for the POU class.
∘ POU5F1P2 [GeneID: 100009665], POU5F1P3 (formerly POUF51L) [GeneID: 5461], POU5F1P4 [GeneID: 100009666], POU5F1P5 [GeneID: 100009667], POU5F1P6 [GeneID: 100009668], POU5F1P7 [GeneID: 100009669] and POU5F1P8 [GeneID: 100009670]. Prior to this study, a single retrotransposed pseudogene of the POU5F1 gene had been annotated and designated POU5F1P1 [Entrez Gene ID: 5462]. Another POU5F1-related sequence of unknown status had been annotated and designated POUF5F1L [GeneID: 5461]. We replace the gene symbol POUF5F1L with POU5F1P3 as this sequence is a retrotransposed pseudogene of POUF51. Our analyses of the human genome sequence identified a further six pseudogenes of POU5F1, which we name sequentially POU5F1P2, POU5F1P4 through to POU5F1P8. Each clearly aligns to the mRNA sequence of POU5F1 but with sequence alterations, indicating origin by retrotransposition. POU5F1P2 and POU5F1P6 have frameshift mutations in the homeobox. POU5F1P5 and POU5F1P6 have stop codons in the homeobox. POU5F1P7 and POU5F1P8 are partial integrants of POU5F1 mRNA excluding the homeobox – POU5F1P7 covers part of the 3' untranslated region and POU5F1P8 a short region around the start codon.
The HNF homeobox class
The HNF class (named after the rat gene Hnf1) encodes proteins with a POU-like domain N-terminal to a highly atypical homeodomain. The POU-like domain, as its name indicates, is weakly similar in sequence to the POU-specific domain [64]; more importantly, it has nearly the same three-dimensional structure and mode of DNA binding as the POU-specific domain [65].
We have identified a total of three HNF-class homeobox genes in the human genome (Tables 1 and 4), consistent with previous work [66,67]. The homeodomains encoded by the human HNF1A and HNF1B genes are atypical in possessing 21 extra amino acid residues between the second and third alpha helices (Additional file 6). We place these two genes in a single gene family (Hnf1) within the HNF class, implying derivation from a single invertebrate gene. Examination of their chromosomal locations concurs with this view. HNF1A and HNF1B map to parts of the genome known to have duplicated in early vertebrate evolution, namely 12q24.31 (HNF1A, near LHX5 and on the same arm as the HOXC cluster) and 17q12 (HNF1B, between LHX1 and the HOXB cluster) (Figure 4). The use of the A and B suffixes is unfortunate, as numerals are generally used to distinguish paralogs of this age, but is retained at present due to widespread and stable use. The homeodomain encoded by the human HMBOX1 gene is atypical in possessing 15 extra amino acid residues between the second and third alpha helices (Additional file 6). Phylogenetic analyses confirm previous suggestions [67] that HMBOX1 is more distantly related to HNF1A and HNF1B (Figure 3; Additional files 1, 2 and 5). We place this gene in a separate gene family (Hmbox) within the same class. We have not found any human HNF-class pseudogenes.
The SINE homeobox class
The SINE class (named after the Drosophila gene so: sine oculis) encodes proteins with a SIX domain N-terminal to a typical homeodomain. The SIX domain is a DNA-binding domain of approximately 115 amino acids; both the SIX domain and the homeodomain are required for DNA binding [68].
We have identified a total of six SINE-class homeobox genes in the human genome (Tables 1 and 4), consistent with previous work [68,69]. The genes form a distinct grouping even if the SIX domain is disregarded – phylogenetic analyses of homeodomains recover the SIX class as a monophyletic group (Figure 3; Additional files 1, 2 and 5). Phylogenetic analyses of homeodomains divide the SIX class into three gene families (Figure 3; Additional files 1, 2 and 5), consistent with previous studies [68,69]. Each gene family has two human members and dates to a single ancestral gene in the most recent common ancestor of bilaterians [68,69]. We have not found any human SINE-class pseudogenes.
The TALE homeobox class
TALE (three amino acid loop extension) class genes are distinguished by the presence of three extra amino acids between the first and second alpha helices of the encoded homeodomain [1,2,70]. Genes belonging to the TALE class encode proteins with various domains outside of the atypical homeodomain.
We have identified a total of 20 TALE-class homeobox genes in the human genome (Tables 1 and 4). The genes form a distinct grouping in phylogenetic analyses even when the three extra homeodomain residues are excluded from the sequence alignment (Figure 3; Additional file 5). Bürglin [2] has given the TALE group the rank of 'superclass' and distinguished between several 'classes' by the presence of distinct domains outside of the homeodomain. These are the IRX domain, MKX domains, the MEIS domain, the PBC domain and TGIF domains [2,71-73]. Along with some others [4,7,24], we have given the TALE group the rank of 'class' containing several 'gene families'; this maintains consistent terminology throughout the present paper. Phylogenetic analyses of homeodomains divide the TALE class into six gene families (Figure 3; Additional files 1, 2 and 5), including an Mkx family containing the recently described MKX gene, which is distinguished from Irx-family genes phylogenetically and by absence of an IRX domain [73,74]. It should be noted that the established name of the Pknox gene family does not indicate orthology with Knox-family genes of plants. We have also identified a total of 10 TALE-class pseudogenes in the human genome (Tables 1 and 4); we have named six of these (IRX4P1, TGIF1P1 and TGIF2P1 to TGIF2P4), and revised the nomenclature of two others (IRX1P1 and PBX2P1).
∘ IRX1P1 [Entrez Gene ID: 646390]. This sequence was previously known as IRXA1; we rename it IRX1P1 because it is clearly a retrotransposed pseudogene of IRX1 and not a functional gene. The IRX1P1 sequence aligns to the mRNA of IRX1 but has a frameshift mutation and two stop codons in the homeobox.
∘ IRX4P1 [Entrez Gene ID: 100009671]. We designate this previously unannotated sequence IRX4P1 because it is clearly a retrotransposed pseudogene of IRX4. The IRX4P1 sequence is a partial integrant derived from a region of the IRX4 mRNA around the stop codon; it lacks the homeobox.
∘ PBX2P1 [Entrez Gene ID: 5088]. This sequence was previously known as PBXP1; we rename it PBX2P1 because it is clearly a retrotransposed pseudogene of PBX2. The former name of PBXP1 did not indicate its transcript of origin. The PBX2P1 sequence aligns to the mRNA of PBX2 but has a frameshift mutation in the coding region.
∘ TGIF1P1 [Entrez Gene ID: 126052]. We designate this previously unannotated sequence TGIF1P1 because it is clearly a retrotransposed pseudogene of TGIF1. The locus has many sequence alterations when compared to TGIF1 mRNA, including a 48 nucleotide insertion within the homeobox.
∘ TGIF2P1 [GeneID: 126826], TGIF2P2 [GeneID: 100009674], TGIF2P3 [GeneID: 100009672] and TGIF2P4 [GeneID: 100009673]. These four sequences were unannotated prior to this study. We designate them TGIF2P1 to TGIF2P4 because they are clearly pseudogenes of TGIF2. Each aligns to the mRNA sequence of TGIF2 but with sequence alterations, indicating origin by retrotransposition. TGIF2P1 has many sequence alterations, including a frameshift mutation in the homeobox. TGIF2P2 and TGIF2P3 are very similar neighboring loci that must have originated by tandem duplication of a retrotransposed TGIF2 mRNA; neither includes the homeobox. TGIF2P4 is a short partial integrant derived from part of the 3' untranslated region of TGIF2 mRNA.
The CUT homeobox class
The CUT class (named after the Drosophila gene cut) generally encodes proteins with one or more CUT domains N-terminal to a typical homeodomain. The CUT domain is a DNA-binding domain of approximately 75 amino acids [75]. There are three widely recognized gene families within the CUT class in humans (Onecut, Cux, Satb; [76]). A fourth gene family (Cmp), lacking a CUT domain but sharing a CMP domain with the Satb gene family, is absent from vertebrates. Bürglin and Cassata [76] have proposed that the vertebrate Satb gene family evolved from the invertebrate Cmp gene family.
We have identified a total of seven CUT-class homeobox genes in the human genome (Tables 1 and 4). Although grouped together by presence of CUT domains, the homeodomains of the Onecut, Cux and Satb gene families are quite divergent and do not always form a monophyletic group in phylogenetic analyses (Additional files 2 and 5). Topologies that separate the gene families are also only weakly supported, so it is most parsimonious to assume that the class is actually monophyletic but the constituent genes underwent rapid sequence divergence following their initial duplications. We have revised the nomenclature of two CUT-class genes (CUX1 and CUX2). We have also identified a total of three CUT-class pseudogenes in the human genome (Tables 1 and 4); we have named all of these (CUX2P1, CUX2P2 and SATB1P1).
∘ CUX1 [Entrez Gene ID: 1523] and CUX2 [Entrez Gene ID: 23316]. These genes were previously known as CUTL1 and CUTL2 respectively. We rename them CUX1 and CUX2 in accordance with homeobox gene nomenclature convention.
∘ CUX2P1 and CUX2P2. These sequences were unannotated prior to this study. We designate them CUX2P1 and CUX2P2 because they are clearly retrotransposed pseudogenes of CUX2. Both are short partial integrants derived from CUX2 mRNA, excluding the homeobox – CUX2P1 covers part of the coding region at the 5' end and CUX2P2 part of the 3' untranslated region.
∘ SATB1P1 [Entrez Gene ID: 100033410]. We designate this previously unannotated sequence SATB1P1 because it is clearly a retrotransposed pseudogene of SATB1. SATB1P1 is a short partial integrant derived from part of the 3' untranslated region of SATB1 mRNA; it does not encompass the homeobox.
The PROS homeobox class
The PROS class (named after the Drosophila gene pros) encodes proteins with a PROS domain C-terminal to an atypical homeodomain. The PROS domain is a DNA-binding domain of approximately 100 amino acids [77]. PROS-class genes encode a highly divergent homeodomain with three extra amino acids. These additional residues are inserted at a different position compared to the TALE class, being between the second and third alpha helices (Additional file 6).
We have identified a total of two PROS-class homeobox genes in the human genome (Tables 1 and 4), which we have placed in a single gene family (Prox). The highly divergent homeodomain sequence and unusual structural features provide justification for PROS being a separate gene class, despite the small number of genes. In phylogenetic analyses, PROS-class homeodomains are situated on a long branch, very distant from other classes (Figure 3; Additional files 1, 2 and 5). The human PROX1 gene is well characterized; we have identified and named its paralog, PROX2. We have not found any human PROS-class pseudogenes.
∘ PROX2 [Entrez Gene ID: 283571]. We designate this previously unannotated gene PROX2 on the basis of clear orthology to the mouse Prox2 gene, inferred from sequence identity and synteny. The homeobox of human PROX2 has two introns and unusually the splice sites of the first (5') intron (AT-AA) do not follow the GT-AG donor-acceptor rule. This has also been noted for mouse Prox2 [78].
The ZF homeobox class
The ZF (zinc finger) class generally encodes proteins with zinc finger motifs, in addition to one or more homeodomains. As noted earlier, phylogenetic analyses of homeodomains does not recover the ZF class as a monophyletic group (Figure 3; Additional files 1, 2 and 5). We recognize that this suggests that zinc finger motifs and homeodomains may have been brought together on three separate occasions in evolution; nonetheless, it is convenient and informative to group these into a single class. Inclusion of the HOMEZ gene in the ZF class may be surprising, as this gene does not encode zinc fingers. However, as previously noted [79] and reproduced in our phylogenetic analyses (Figure 3; Additional files 1, 2 and 5), the multiple homeodomain sequences of this gene are clearly related to those encoded by the ZHX1, ZHX2 and ZHX3 genes.
We have identified a total of 14 ZF-class homeobox genes in the human genome (Tables 1 and 4), which we have placed in five gene families (Adnp, Tshz, Zeb, Zfhx and Zhx/Homez). We have also identified one ZF-class pseudogenes in the human genome (Tables 1 and 4). We have revised the nomenclature of five of these loci (ADNP2, ZEB1, ZEB2, ZEB2P1 and ZFHX3).
∘ ADNP2 [Entrez Gene ID 22850]. This gene was previously known as ZNF508; we rename it ADNP2 to reflect its paralogous relationship to ADNP.
∘ ZEB1 [Entrez Gene ID: 6935] and ZEB2 [Entrez Gene ID: 9839]. These genes were previously known as ZFHX1A and ZFHX1B respectively. We rename them ZEB1 and ZEB2 to distinguish them from genes belonging to the distantly related Zfhx gene family.
∘ ZEB2P1 [Entrez Gene ID: 100033412]. This retrotransposed pseudogene of ZEB2 has been described previously [80]. Our new nomenclature (ZEB2P1) reflects the origin of this locus.
∘ ZFHX3 [Entrez Gene ID: 463]. This gene was previously known as ATBF1; we rename it ZFHX3 to reflect its close relationship to ZFHX2 and ZFHX4; indeed ZFHX3 was a synonym for this gene.
The CERS homeobox class
The highly unusual CERS (ceramide synthase) class, also known as the LASS (longevity assurance) class, comprises a single gene family that is highly conserved amongst eukaryotes and includes the yeast gene and original member LAG1. There are six CERS-class genes in the human genome (CERS1 to CERS6) and five of these (CERS2 to CERS6) encode proteins with a homeodomain sequence [81,82]. These are, however, extremely divergent from the homeodomains of other gene classes. Secondary structure prediction analyses suggest these sequences have the potential to encode three alpha helices in the appropriate positions (data not shown). The most surprising characteristic of these genes is that biochemical studies predict them to encode transmembrane proteins, with the homeodomain on the cytosolic side of the endoplasmic reticulum membrane, and hence they could not act as DNA-binding proteins or transcription factors [81,82]. It is possible that an ancestor of these genes gained a homeobox through exon shuffling, or alternatively this could represent convergent evolution. We include only CERS2 to CERS6 in our comprehensive compilation of human homeobox genes, as CERS1 lacks a homeobox motif.
Chromosomal distribution of human homeobox genes
The chromosomal locations of genes can give clues to evolutionary ancestry, including patterns of gene duplication, and the possible existence of gene clusters. In Figure 4, we show the chromosomal locations of all human homeobox genes. We do not include probable pseudogenes on these ideograms, because most of these have originated by reverse transcription of mRNA and secondary integration into the genome, and hence give no insight into ancestral locations of genes. The highly repetitive DUX1 to DUX5 sequences are also not shown, as these have undergone secondary amplification and are also most likely non-functional (see above).
The first observation is that there are homeobox genes on every human chromosome. Even the two sex chromosomes harbor homeobox genes, with SHOX (short stature homeobox) in the PAR1 pseudoautosomal region at the tip of the short arms of X and Y being the best known. Haploinsufficiency of SHOX is implicated in the short stature phenotype of Turner syndrome patients who lack one copy of the X chromosome [83]. There are also nine other homeobox genes in non-pseudoautosomal regions of the X chromosome, including three tandemly-arranged members of the Rhox gene family, collectively homologous to the multiple Rhox (reproductive homeobox) genes of mouse. Only one of the homeobox genes on the X chromosome, the TALE-class gene TGIF2LX, has a distinct homolog on the Y chromosome, called TGIF2LY. These genes map to the largest homology block shared by the unique regions of the X and Y chromosomes, spanning 3.5 Mb. It has been proposed that the ancestor of these two genes arose by retrotransposition of TGIF2 mRNA [84].
The autosomes with the lowest number of homeobox genes are chromosomes 21 (with just PKNOX1) and 22 (with GSC2 and ISX). Examination of the remaining autosomes reveals that homeobox genes are quite dispersed with some interesting regional accumulations. The best known examples of close linkage between homeobox genes are the four Hox clusters on human chromosomes 2, 7, 12 and 17, comprising 9, 11, 9 and 10 genes respectively; each of these is shown as just a single line on each ideogram for simplicity (Figure 4). These should not be considered in isolation, however, because many other ANTP-class genes map in the vicinity of the Hox clusters [26,27]. These include genes very tightly linked to the Hox clusters, notably the Evx-family genes (on chromosomes 2 and 7), Dlx-family genes (on chromosomes 2 and 17), and Meox-family genes (on chromosomes 2 and 17).
There are other concentrations of ANTP-class genes away from the Hox clusters. These are the ParaHox cluster (GSX1, PDX1, CDX2) on chromosome 13, and four sets of NKL-subclass genes on 2p/8p (split), 4p, 5q and 10q, hypothesized to be derived from an ancestral array by duplication [26,33]. The accumulation on the distal half of the long arm of chromosome 10 is particularly striking, comprising eleven ANTP-class genes from 10 gene families. This is not a tight gene cluster, but it is compatible with ancestry by extensive tandem gene duplication followed by dispersal. Discounting the rather aberrant case of the Hox clusters, this region of the long arm of chromosome 10 is the most homeobox-rich region of the human genome.
There are additional groupings of homeobox genes outside the ANTP class. These include two TALE-class Irx clusters on chromosomes 5 and 16 homologous to the described mouse Irx clusters [19], and a set of PRD-class genes on chromosome 19 proposed to be derived from the CRX homeobox gene by duplication and rapid divergence [21]. Perhaps the most interesting case, however, is found on the tip of the long arm of chromosome 9, where there is a concentration of homeobox genes from disparate gene classes. Four LIM-class genes, one ANTP-class gene, one PRD-class gene and one TALE-class gene are found in this location. Although dispersed over a large region, and not forming a tight gene cluster, the linkages are nonetheless intriguing. It is possible that these linkages reflect ancestry from the very ancient gene duplications that must have generated the distinctive homeobox gene classes found within animal genomes.
Conclusion
We identified 300 homeobox loci in the euchromatic regions of the human genome, and divide these into 235 probable functional genes and 65 probable pseudogenes. Not all of these loci possess a homeobox because for completeness we include all sequences derived from homeobox-containing genes. The number of homeobox sequences is also different from the number of loci because several genes contain multiple homeobox motifs. The figures exclude the repetitive DUX1 to DUX5 homeobox sequences of which we identified 35 probable pseudogenes, with many more expected in heterochromatic regions.
New or revised nomenclature is proposed for approximately 70 of the 300 homeobox loci in order to clarify orthologous relationships between human and mouse, to indicate evolutionary relationships within a gene family, to distinguish genes from pseudogenes, and to indicate pseudogene origins. The loci are also classified into a simple hierarchical scheme, comprising 102 gene families within eleven gene classes. The classification scheme proposed may be widely applicable to homeobox genes from other animals.
The 235 probable functional homeobox genes map to every human chromosome with some interesting regional concentrations of genes. These include a large number of ANTP-class genes on the distal end of the long arm of chromosome 10, and a combination of LIM-, ANTP-, PRD- and TALE-class genes on the distal end of the long arm of chromosome 9. These associations may be remnants of common ancestry early in animal evolution.
Methods
The finished human genome sequence (build 35.1) was subjected to a series of tBLASTn searches [85,86] using known homeodomain sequences from the ANTP, PRD, LIM, POU, HNF, SINE, TALE, CUT, PROS and ZF classes. No arbitrary E-value cut-off was selected, but instead each list of hits was analyzed manually until true homeodomain sequences ceased to be detected. Definition of a homeodomain used a combination of CD-search for conserved protein domains implemented through BLASTp [85,86] and secondary structure prediction by JPred implemented through the Barton Group, University of Dundee [87]. Each time a new or divergent homeodomain match was found, the tBLASTn process was repeated. Six very divergent gene families were undetected by this method but found by text searching: Hopx, Adnp, Tshz, Zeb, Zhx/Homez and Cers. To ensure that every pseudogene was detected, including truncated or decayed versions lacking the homeobox, the full mRNA sequence of each gene was deduced and used in a BLASTn search of the human genome sequence [85,86]. Pseudogenes were recognized as those genomic regions with similarity to non-repetitive DNA sequences of the parent gene, even if aligning to only part of the locus. Pseudogenes undergo mutational decay and would eventually become unrecognizable, but in practice ambiguous cases were not encountered. Exon-intron structures of novel loci were deduced by comparison between genomic sequence and cDNA, EST or retrotransposed pseudogene sequences, as previously described [21]. Several unnamed human loci were identified as probable orthologs of known mouse genes; orthology was deduced by a combination of homeodomain sequence similarity and synteny, examined through the mouse genome sequence (build 34.1) and the Ensembl Genome Browser [88].
Phylogenetic analyses were performed with homeodomain sequences, after each had been edited to an alignment of 60 amino acids (Additional file 7), using the maximum likelihood [89] and neighbor-joining [90] methods. Maximum likelihood trees were constructed using PhyML [91], with a JTT model of amino acid substitution, four categories of between-site rate heterogeneity, a gamma distribution parameter estimated from the data and 500 bootstrap resamplings. Neighbor-joining trees were constructed using PHYLIP (.)[92], with a JTT model of amino acid substitution and 1000 bootstrap resamplings. For defining human gene families, all Drosophila homeodomains were first combined with all human homeodomains in maximum likelihood and neighbor-joining analyses to enable divergent Drosophila genes to be identified and removed. These include genes lost from human, as well genes known to have undergone unusually rapid evolution in Drosophila. For the Hox3 family the rapidly evolving Drosophila genes bcd, zen and zen2 were then replaced by an ortholog from centipede (Sm Hox3b), and for the Nk4 family the rapid evolving Drosophila gene tin was replaced by an ortholog from annelid (Pd NK4). In addition, six genes from other protostome or cnidarian genomes were added to represent gene families known to be missing from Drosophila (Pdx family: Ps Xlox; Alx family: Nv CART1; Dmbx family: Hv manacle; Pou1 family: Nv POU1; Hnf1 family: Nv HNF; Pknox family: Am Prep). Only 100 bootstrap resamplings were performed on this dataset because of its large size (354 homeodomains). Trees were displayed using TreeExplorer [93]. Genes encoding partial homeodomains, and probable pseudogenes, were not included in the phylogenetic analyses. With short alignments, phylogenetic trees can only be used as guides to relationships, not absolute indicators of evolutionary history, and the trees presented in this paper should be interpreted in this light.
Authors' contributions
PWHH designed the study and contributed to gene identification and to gene nomenclature revisions. HAFB carried out database searches, annotations and phylogenetic analyses and contributed to gene nomenclature revisions. PWHH and HAFB drafted the manuscript. EB contributed to gene nomenclature revisions, discussed these with the research community and databases, and implemented the agreed changes.
All authors edited and approved the final manuscript.
Supplementary Material
Maximum likelihood phylogenetic tree of all human plus selected protostome and cnidarian homeodomains for identification of gene families. Arbitrarily rooted phylogenetic tree of all human plus selected protostome and cnidarian homeodomains constructed using the maximum likelihood (ML) method. Bootstrap values supporting gene family designations are shown. Homeodomain sequences derived from pseudogenes are excluded. This ML tree should be compared with the neighbor-joining (NJ) tree shown in Additional file 2. The dataset used for both ML and NJ analyses includes all human homeodomains, most Drosophila melanogaster homeodomains, plus selected additional homeodomains from other protostomes or cnidarians when the gene family is divergent or absent in Drosophila. Divergent Drosophila genes that do not group with human genes were identified by construction of a preliminary, non-bootstrapped ML and NJ trees, and subsequently removed from the dataset. These include genes lost from human, as well genes known to have undergone unusually rapid evolution in Drosophila. For the Hox3 family the rapidly evolving Drosophila genes bcd, zen and zen2 were replaced with Sm Hox3b, and for the Nk4 family the rapid evolving Drosophila gene tin was replaced with Pd NK4. In addition, six genes from other protostome or cnidarian genomes were added to represent gene families known to be missing from Drosophila (Pdx family: Ps Xlox; Alx family: Nv CART1; Dmbx family: Hv manacle; Pou1 family: Nv POU1; Hnf1 family: Nv HNF; Pknox family: Am Prep). Species abbreviations: Am, Apis mellifera (honeybee); Dm, Drosophila melanogaster (fruitfly); Hv, Hydra vulgaris (hydrozoan); Nv, Nematostella vectensis (starlet sea anemone); Pd, Platynereis dumerilii (annelid worm); Ps, Phascolion strombus (sipunculan worm); Sm, Strigamia maritima (centipede). ML performed more poorly than NJ in recovering several well known gene families, notably Hox4, Hox5, Nk4 and Alx. In contrast, ML did recover PROP1 and CG32532 as a true gene family; NJ did not. The invertebrate gene does not always lie as a strict outgroup to all human genes in a family; this effect is expected when using a short alignment. Instead, distinct grouping of invertebrate and human genes is taken as evidence of ancestry from a single gene. A few ambiguous cases were encountered, notably divergence of Drosophila H2.0 in the proposed Hlx gene family, and resolution within the Pax4/6 gene family, which is recovered as two families in NJ but one in ML. As explained in the text, several human gene families contain 'orphan' genes without invertebrate orthologs; these are Barx, Nanog, Noto, Vax, Ventx, Argfx, Dprx, Dux, Esx, Hesx, Hopx, Isx, Leutx, Mix, Nobox, Rhox, Sebox, Tprx, Hdx, Pou5, Hmbox, Satb, Adnp and Zhx/Homez. Zeb and Mkx would be placed in this category based on our ML and NJ trees, although other data suggest that Drosophila zfh1 and CG11617 respectively may be the protostome orthologs [73,94]. Tshz is only an apparent orphan family; the clear Drosophila ortholog simply lacks the homeobox [95,96]. Phylogenetic analysis is just one source of evidence for allocation of genes to gene families and identification of boundaries between gene families; complementary criteria used are synteny between species and paralogy within the human genome. Our ML and NJ trees should not be used to allocate gene families to gene classes, because other diagnostic characters such as insertions within the homeodomain, key amino acid residues, and several motifs outside of the homeodomain are excluded from the analysis. Indeed, artefactual mixing of the TALE and SINE classes occurs in both ML and NJ trees.
Neighbor-joining phylogenetic tree of all human plus selected protostome and cnidarian homeodomains for identification of gene families. Arbitrarily rooted phylogenetic tree of all human plus selected protostome and cnidarian homeodomains constructed using the neighbor-joining (NJ) method. Bootstrap values supporting gene family designations are shown. Homeodomain sequences derived from pseudogenes are excluded. Comparison of NJ and ML trees, and description of the dataset used, is given in the legend to Additional file 1. Several artefactual mixing of classes occurs in this NJ tree, notably splitting of the CUT class, mixing of the TALE and SINE classes and aberrant placement of HOPX.
Neighbor-joining phylogenetic tree of human ANTP-class homeodomains, for comparison to maximum likelihood tree. Arbitrarily rooted phylogenetic tree of human ANTP-class homeodomains constructed using the neighbor-joining method. Bootstrap values supporting internal nodes with over 70% are shown. Homeodomain sequences derived from pseudogenes are excluded. The proposed division between the HOXL and NKL subclasses is indicated. The position of EN1 and EN2 is unstable; this tree places them close to the base of the HOXL/NKL divergence, whereas maximum likelihood analysis of the same dataset places them firmly in the NKL subclass (Figure 1). Interrelationships of genes in the Nk2.2 and Nk4 families are also unstable (in this tree and Figure 1 respectively); in these cases synteny within and between genomes clearly resolves gene families. Detailed relationships between different gene families should not be inferred from this tree.
Neighbor-joining phylogenetic tree of human PRD-class homeodomains, for comparison to maximum likelihood tree. Arbitrarily rooted phylogenetic tree of human PRD-class homeodomains constructed using the neighbor-joining method. Bootstrap values supporting internal nodes with over 70% are shown. Homeodomain sequences derived from pseudogenes are excluded, as are the partial homeodomains of PAX2, PAX5 and PAX8, and the HOPX homeodomain because its extremely divergent sequence destabilizes the overall tree topology. Roman numeral suffixes are used to distinguish multiple homeodomains encoded by a single Dux-family gene. Detailed relationships between different gene families should not be inferred from this tree.
Neighbor-joining phylogenetic tree of human homeodomains excluding ANTP and PRD classes, for comparison to maximum likelihood tree. Arbitrarily rooted phylogenetic tree of human homeodomains excluding the ANTP and PRD classes constructed using the neighbor-joining method. Bootstrap values supporting internal nodes with over 70% are shown. Homeodomain sequences derived from pseudogenes are excluded. Roman numeral suffixes are used to distinguish multiple homeodomains encoded by a single gene. Classes and/or families are color coded as shown in the key. The LIM and ZF classes are not recovered as two distinct monophyletic groups, a result also found by maximum likelihood analysis (Figure 3). The multiple homeodomains of Zfhx-family proteins and Zhx/Homez-family proteins are also dispersed in the tree, presumably artefactually. Monophyly of the CUT class is not recovered in this tree, but is by maximum likelihood analysis (Figure 3). Detailed relationships between different gene families should not be inferred from this tree.
Multiple sequence alignment of all human plus selected protostome and cnidarian homeodomains. The consensus homeodomain sequence (shown several times for reference) was derived from a compilation of 247 human homeodomain sequences. The three horizontal lines indicate the positions of the three alpha-helices. The numbering scheme refers to amino acid position in the canonical 60-amino-acid homeodomain; insertions relative to this sequence are shown when present. Black shaded resides are invariant between all human homeodomains within each class (or family in the case of the ZF homeodomains). Sequence accession numbers are shown. For each gene family designation, maximum likelihood and neighbor-joining bootstrap support values are indicated (see Additional files 1 and 2). These values are not shown if the gene family does not form a monophyletic group in phylogenetic analyses (in which case n/a is written) or if an invertebrate homolog could not be found.
Phylogenetic input file. All human and invertebrate homeodomains used in phylogenetic analyses are shown, after alignment and removal of insertions to give a uniform 60-amino-acid alignment.
Acknowledgments
Acknowledgements
We thank Rebecca Furlong, Tokiharu Takahashi, Hidetoshi Saiga, Naohito Takatori, David Ferrier, Mario Pestarino, Thomas Bürglin and reviewers for helpful advice. Research undertaken by PWHH and HAFB was supported by the BBSRC and the Wellcome Trust. The work of EAB and the HUGO Gene Nomenclature Committee is supported by NHGRI grant P41 HG003345 and the Wellcome Trust.
Contributor Information
Peter WH Holland, Email: peter.holland@zoo.ox.ac.uk.
H Anne F Booth, Email: anne.booth@merton.oxon.org.
Elspeth A Bruford, Email: hgnc@genenames.org.
References
- Bürglin TR. A comprehensive classification of homeobox genes. In: Duboule D, editor. Guidebook to the Homeobox Genes. Oxford: Oxford University Press; 1994. pp. 25–71. [Google Scholar]
- Bürglin TR. Homeodomain proteins. In: Meyers RA, editor. Encyclopedia of Molecular Cell Biology and Molecular Medicine. 2. Vol. 6. Weinheim: Wiley-VCH Verlag GmbH & Co; 2005. pp. 179–222. [Google Scholar]
- Boncinelli E. Homeobox genes and disease. Curr Op Genet Dev. 1997;7:331–337. doi: 10.1016/S0959-437X(97)80146-3. [DOI] [PubMed] [Google Scholar]
- Edvardsen RB, Seo H-C, Jensen MF, Mialon A, Mikhaleva J, Bjordal M, Cartry J, Reinhardt R, Weissenbach J, Wincker P, et al. Remodelling of the homeobox gene complement in the tunicate Oikopleura dioica. Curr Biol. 2005;15:R12–R13. doi: 10.1016/j.cub.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Galliot B, de Vargas C, Miller D. Evolution of homeobox genes: Q50 Paired-like genes founded the Paired class. Dev Genes Evol. 1999;209:186–197. doi: 10.1007/s004270050243. [DOI] [PubMed] [Google Scholar]
- Holland PWH, Takahashi T. The evolution of homeobox genes: implications for the study of brain development. Brain Res Bull. 2005;66:484–490. doi: 10.1016/j.brainresbull.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Ryan JF, Burton PM, Mazza ME, Kwong GK, Mullikin JC, Finnerty JR. The cnidarian-bilaterian ancestor possessed at least 56 homeoboxes: evidence from the starlet sea anemone, Nematostella vectensis. Genome Biol. 2006;7:R64. doi: 10.1186/gb-2006-7-7-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee-Basu S, Baxevanis AD. Molecular evolution of the homeodomain family of transcription factors. Nucleic Acids Res. 2001;29:3258–3269. doi: 10.1093/nar/29.15.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- IHGSC Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Nam J, Nei M. Evolutionary change of the numbers of homeobox genes in bilateral animals. Mo Bio Evol. 2005;22:2386–2394. doi: 10.1093/molbev/msi229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckers M-C, Gabriëls J, van der Maarel S, De Vriese A, Frants RR, Collen D, Belayew A. Active genes in junk DNA? Characterization of DUX genes embedded within 3.3 kb repeated elements. Gene. 2001;264:51–57. doi: 10.1016/S0378-1119(00)00602-8. [DOI] [PubMed] [Google Scholar]
- Ding H, Beckers M-C, Plaisance S, Marynen P, Collen D, Belayew A. Characterization of a double homeodomain protein (DUX1) encoded by a cDNA homologous to 3.3 kb dispersed repeated elements. Hum Mol Genet. 1998;7:1681–1694. doi: 10.1093/hmg/7.11.1681. [DOI] [PubMed] [Google Scholar]
- Gabriëls J, Beckers M-C, Ding H, De Vriese A, Plaisance S, van der Maarel SM, Padberg GW, Frants RR, Hewitt JE, Collen D, et al. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999;236:25–32. doi: 10.1016/S0378-1119(99)00267-X. [DOI] [PubMed] [Google Scholar]
- Akam ME, Holland PWH, Ingham PW, Wray G. The evolution of developmental mechanisms. Development. 1994. pp. 135–142.
- Joyner AL, Hanks M. The engrailed genes: evolution of function. Semin Dev Bio. 1991;2:435–445. [Google Scholar]
- Echelard Y, Epstein DJ, St-Jacques B, Shen L, Mohler J, McMahon JA, McMahon AP. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75:1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- Stock DW, Ellies DL, Zhao Z, Ekker M, Ruddle FH, Weiss KM. The evolution of the vertebrate Dlx genefamily. Proc Natl Acad Sci USA. 1996;93:10858–10863. doi: 10.1073/pnas.93.20.10858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters T, Dildrop R, Ausmeier K, Ruther U. Organization of mouse Iroquois homeobox genes in two clusters suggests a conserved regulation and function in vertebrate development. Genome Res. 2000;10:1453–1462. doi: 10.1101/gr.144100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rosa R, Grenier JK, Andreeva T, Cook CE, Adoutte A, Akam M, Carroll SB, Balavoine G. Hox genes in brachiopods and priapulids and protostome evolution. Nature. 1999;399:772–776. doi: 10.1038/21631. [DOI] [PubMed] [Google Scholar]
- Booth HAF, Holland PWH. Annotation, nomenclature and evolution of four novel homeobox genes expressed in the human germ line. Gene. 2007;387:7–14. doi: 10.1016/j.gene.2006.07.034. [DOI] [PubMed] [Google Scholar]
- Booth HAF, Holland PWH. Eleven daughters of NANOG. Genomics. 2004;84:229–238. doi: 10.1016/j.ygeno.2004.02.014. [DOI] [PubMed] [Google Scholar]
- Castro LFC, Rasmussen SLK, Holland PWH, Holland ND, Holland LZ. A Gbx homeobox gene in amphioxus: insights into ancestry of the ANTP class and evolution of the midbrain/hindbrain boundary. Dev Biol. 2006;295:40–51. doi: 10.1016/j.ydbio.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Dearden PK, Wilson MJ, Sablan L, Osborne PW, Havler M, McNaughton E, Kimura K, Milshina NV, Hasselmann M, Gempe T, et al. Patterns of conservation and change in honey bee developmental genes. Genome Res. 2006;16:1376–1384. doi: 10.1101/gr.5108606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro AS, Schierwater B, Dellaporta SL, Holland PWH. A low diversity of ANTP class homeobox genes in Placozoa. Evol Dev. 2006;8:174–182. doi: 10.1111/j.1525-142X.2006.00087.x. [DOI] [PubMed] [Google Scholar]
- Castro LFC, Holland PWH. Chromosomal mapping of ANTP class homeobox genes in amphioxus: piecing together ancestral genomes. Evol Dev. 2003;5:459–465. doi: 10.1046/j.1525-142X.2003.03052.x. [DOI] [PubMed] [Google Scholar]
- Pollard SL, Holland PWH. Evidence for 14 homeobox gene clusters in human genome ancestry. Curr Biol. 2000;10:1059–1062. doi: 10.1016/S0960-9822(00)00676-X. [DOI] [PubMed] [Google Scholar]
- Brooke NM, Garcia-Fernàndez J, Holland PWH. The ParaHox gene cluster is an evolutionary sister of the Hox gene cluster. Nature. 1998;392:920–922. doi: 10.1038/31933. [DOI] [PubMed] [Google Scholar]
- Ferrier DEK, Brooke NM, Panopoulou G, Holland PWH. The Mnx homeobox gene class defined by HB9, MNR2 and amphioxus AmphiMnx. Dev Genes Evol. 2001;211:103–107. doi: 10.1007/s004270000124. [DOI] [PubMed] [Google Scholar]
- Venkatesh TV, Holland ND, Holland LZ, Su M-T, Bodmer R. Sequence and developmental expression of amphioxus AmphiNk2-1: insights into the evolutionary origin of the vertebrate thyroid gland and forebrain. Dev Genes Evol. 1999;209:254–259. doi: 10.1007/s004270050250. [DOI] [PubMed] [Google Scholar]
- Holland ND, Venkatesh TV, Holland LZ, Jacobs DK, Bodmer R. Amphink2-tin, an amphioxus homeobox gene expressed in myocardial progenitors: insights into evolution of the vertebrate heart. Dev Biol. 2003;255:128–137. doi: 10.1016/S0012-1606(02)00050-7. [DOI] [PubMed] [Google Scholar]
- Hislop NR, de Jong D, Hayward DC, Ball EE, Miller DJ. Tandem organization of independently duplicated homeobox genes in the basal cnidarian Acropora millepora. Dev Genes Evol. 2005;215:268–273. doi: 10.1007/s00427-005-0468-y. [DOI] [PubMed] [Google Scholar]
- Luke GN, Castro LFC, McLay K, Bird C, Coulson A, Holland PWH. Dispersal of NK homeobox gene clusters in amphioxus and humans. Proc Natl Acad Sci USA. 2003;100:5292–5295. doi: 10.1073/pnas.0836141100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiojima I, Komuro I, Mizuno T, Aikawa R, Akazawa H, Oka T, Yamazaki T, Yazaki Y. Molecular cloning and characterization of human cardiac homeobox gene CSX1. Circulation Res. 1996;79:920–929. doi: 10.1161/01.res.79.5.920. [DOI] [PubMed] [Google Scholar]
- Moretti P, Simmons P, Thomas P, Haylock D, Rathjen P, Vadas M, D'Andrea R. Identification of homeobox genes expressed in human haemopoietic progenitor cells. Gene. 1994;144:213–219. doi: 10.1016/0378-1119(94)90380-8. [DOI] [PubMed] [Google Scholar]
- Hart AH, Hartley L, Ibrahim M, Robb L. Identification, cloning and expression analysis of the pluripotency promoting Nanog genes in mouse and human. Dev Dynamics. 2004;230:187–198. doi: 10.1002/dvdy.20034. [DOI] [PubMed] [Google Scholar]
- Fairbanks D, Maughan P. Evolution of the NANOG pseudogene family in the human and chimpanzee genomes. BMC Evol Biol. 2006;6:12. doi: 10.1186/1471-2148-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wang X, Li M, Han J, Chen B, Wang B, Dai J. NANOGP8 is a retrogene expressed in cancers. FEBS J. 2006;273:1723–1730. doi: 10.1111/j.1742-4658.2006.05186.x. [DOI] [PubMed] [Google Scholar]
- Moreau-Aubry A, Le Guiner S, Labarrière N, Gesnel M-C, Jotereau F, Breathnach R. A processed pseudogene codes for a new antigen recognized by a CD8+ T cell clone on melanoma. J Exp Med. 2000;191:1617–1623. doi: 10.1084/jem.191.9.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balczarek KA, Lai Z-C, Kumar S. Evolution and functional diversification of the paired box (Pax) DNA-binding domains. Mol Biol Evol. 1997;14:829–842. doi: 10.1093/oxfordjournals.molbev.a025824. [DOI] [PubMed] [Google Scholar]
- Chen F, Kook H, Milewski R, Gitler AD, Lu MM, Li J, Nazarian R, Schnepp R, Jen K, Biben C, et al. Hop is an unusual homeobox gene that modulates cardiac development. Cell. 2002;110:713–723. doi: 10.1016/S0092-8674(02)00932-7. [DOI] [PubMed] [Google Scholar]
- Shin CH, Liu Z-P, Passier R, Zhang C-L, Wang D-Z, Harris TM, Yamagishi H, Richardson JA, Childs G, Olson EN. Modulation of cardiac growth and development by HOP, an unusual homeodomain protein. Cell. 2002;110:725–735. doi: 10.1016/S0092-8674(02)00933-9. [DOI] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Gulisano M, Stornaiuolo A, Boncinelli E. Nested expression domains of four homeobox genes in developing rostral brain. Nature. 1992;358:687–690. doi: 10.1038/358687a0. [DOI] [PubMed] [Google Scholar]
- Plouhinec J-L, Sauka-Spengler T, Germot A, Le Mentec C, Cabana T, Harrison G, Pieau C, Sire J-Y, Véron G, Mazan S. The mammalian Crx genes are highly divergent representatives of the Otx5 gene family, a gnathostome orthology class of orthodenticle-related homeogenes involved in the differentiation of retinal photoreceptors and circadian entrainment. Mol Biol Evol. 2003;20:513–521. doi: 10.1093/molbev/msg085. [DOI] [PubMed] [Google Scholar]
- Wada H, Saiga H, Satoh N, Holland PWH. Tripartite organization of the ancestral chordate brain and the antiquity of placodes: insights from ascidian Pax-2/5/8, Hox and Otx genes. Development. 1998;125:1113–1122. doi: 10.1242/dev.125.6.1113. [DOI] [PubMed] [Google Scholar]
- Wada H, Holland PWH, Sato S, Yamamoto H, Satoh N. Neural tube is partially dorsalized by overexpression of HrPax-37: the ascidian homologue of Pax-3 and Pax-7. Dev Biol. 1997;187:240–252. doi: 10.1006/dbio.1997.8626. [DOI] [PubMed] [Google Scholar]
- MacLean JA, 2nd, Chen MA, Wayne CM, Bruce SR, Rao M, Meistrich ML, Macleod C, Wilkinson MF. Rhox: a new homeobox gene cluster. Cell. 2005;120:369–382. doi: 10.1016/j.cell.2004.12.022. [DOI] [PubMed] [Google Scholar]
- Jackson M, Watt AJ, Gautier P, Gilchrist D, Driehaus J, Graham GJ, Keebler J, Prugnolle F, Awadalla P, Forrester LM. A murine specific expansion of the Rhox cluster involved in embryonic stem cell biology is under natural selection. BMC Genom. 2006;7:212. doi: 10.1186/1471-2164-7-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean JA, 2nd, Lorenzetti D, Hu Z, Salerno WJ, Miller J, Wilkinson MF. Rhox homeobox gene cluster: recent duplication of three family members. Genesis. 2006;44:122–129. doi: 10.1002/gene.20193. [DOI] [PubMed] [Google Scholar]
- Morris L, Gordon J, Blackburn CC. Identification of a tandem duplicated array in the Rhox alpha locus on mouse chromosome X. Mamm Genome. 2006;17:178–187. doi: 10.1007/s00335-005-0138-4. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang J. Remarkable expansions of an X-linked reproductive homeobox gene cluster in rodent evolution. Genomics. 2006;88:34–43. doi: 10.1016/j.ygeno.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Wimmer K, Zhu X-X, Rouillard JM, Ambros PF, Lamb BJ, Kuick R, Eckart M, Weinhäusl A, Fonatsch C, Hanash SM. Combined restriction landmark genomic scanning and virtual genome scans identify a novel human homeobox gene, ALX3, that is hypermethylated in neuroblastoma. Genes Chromosomes Cancer. 2002;33:285–294. doi: 10.1002/gcc.10030. [DOI] [PubMed] [Google Scholar]
- Norris RA, Scott KK, Moore CS, Stetten G, Brown CR, Jabs EW, Wulfsberg EA, Yu J, Kern MJ. Human PRRX1 and PRRX2 genes: cloning, expression, genomic localization, and exclusion as disease genes for Nager syndrome. Mamm Genome. 2000;11:1000–1005. doi: 10.1007/s003350010193. [DOI] [PubMed] [Google Scholar]
- Saito T, Greenwood A, Sun Q, Anderson DJ. Identification by differential RT-PCR of a novel paired homeodomain protein specifically expressed in sensory neurons and a subset of their CNS targets. Mol Cell Neurosci. 1995;6:280–292. doi: 10.1006/mcne.1995.1022. [DOI] [PubMed] [Google Scholar]
- Heathcote K, Braybrook C, Abushaban L, Guy M, Khetyar ME, Patton MA, Carter ND, Scambler PJ, Syrris P. Common arterial trunk associated with a homeodomain mutation of NKX2.6. Hum Mol Genet. 2005;14:585–593. doi: 10.1093/hmg/ddi055. [DOI] [PubMed] [Google Scholar]
- Wayne CM, MacLean JA, 2nd, Cornwall G, Wilkinson MF. Two novel human X-linked homeobox genes, hPEPP1 and hPEPP2, selectively expressed in the testis. Gene. 2002;301:1–11. doi: 10.1016/S0378-1119(02)01087-9. [DOI] [PubMed] [Google Scholar]
- Cinquanta M, Rovescalli AC, Kozak CA, Nirenberg M. Mouse Sebox homeobox gene expression in skin, brain, oocytes, and two-cell embryos. Proc Natl Acad Sci USA. 2000;97:8904–8909. doi: 10.1073/pnas.97.16.8904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijmenga C, Frants RR, Hewitt JE, van Deutekom JCT, van Geel M, Wright TJ, Padberg GW, Hofker MH, van Ommen G-JB. Molecular genetics of facioscapulohumeral muscular dystrophy. Neuromusc Dis. 1993;3:487–491. doi: 10.1016/0960-8966(93)90102-P. [DOI] [PubMed] [Google Scholar]
- Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- Hobert O, Westphal H. Functions of LIM-homeobox genes. Trends Genet. 2000;16:75–83. doi: 10.1016/S0168-9525(99)01883-1. [DOI] [PubMed] [Google Scholar]
- Phillips K, Luisi B. The virtuoso of versatility: POU proteins that flex to fit. J Mol Biol. 2000;302:1023–1039. doi: 10.1006/jmbi.2000.4107. [DOI] [PubMed] [Google Scholar]
- Ryan AK, Rosenfeld MG. POU domain family values: flexibility, partnerships, and developmental codes. Genes Dev. 1997;11:1207–1225. doi: 10.1101/gad.11.10.1207. [DOI] [PubMed] [Google Scholar]
- Andersen B, Rosenfeld MG. POU domain factors in the neuroendocrine system: lessons from developmental biology provide insights into human disease. Endocrine Rev. 2001;22:2–35. doi: 10.1210/er.22.1.2. [DOI] [PubMed] [Google Scholar]
- Baumhueter S, Mendel DB, Conley PB, Kuo CJ, Turk C, Graves MK, Edwards CA, Courtois G, Crabtree GR. HNF-1 shares three sequence motifs with the POU domain proteins and is identical to LF-B1 and APF. Genes Dev. 1990;4:372–379. doi: 10.1101/gad.4.3.372. [DOI] [PubMed] [Google Scholar]
- Chi Y-I, Frantz JD, Oh B-C, Hansen L, Dhe-Paganon S, Shoelson SE. Diabetes mutations delineate an atypical POU domain in HNF-1alpha. Mol Cell. 2002;10:1129–1137. doi: 10.1016/S1097-2765(02)00704-9. [DOI] [PubMed] [Google Scholar]
- Bach I, Mattei M-G, Cereghini S, Yaniv M. Two members of an HNF1 homeoprotein family are expressed in human liver. Nucleic Acids Res. 1991;19:3553–3559. doi: 10.1093/nar/19.13.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Saiyin H, Zeng X, Xi J, Liu X, Li X, Yu L. Isolation and functional analysis of human HMBOX1, a homeobox containing protein with transcriptional repressor activity. Cytogen Genome Res. 2006;114:131–136. doi: 10.1159/000093328. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Sato S, Ozaki H, Ikeda K. Six family genes-structure and function as transcription factors and their roles in development. BioEssays. 2000;22:616–626. doi: 10.1002/1521-1878(200007)22:7<616::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Gallardo ME, Lopez-Rios J, Fernaud-Espinosa I, Granadino B, Sanz R, Ramos C, Ayuso C, Seller MJ, Brunner HG, Bovolenta P, et al. Genomic cloning and characterization of the human homeobox gene SIX6 reveals a cluster of SIX genes in chromosome 14 and associates SIX6 hemizygosity with bilateral anophthalmia and pituitary anomalies. Genomics. 1999;61:82–91. doi: 10.1006/geno.1999.5916. [DOI] [PubMed] [Google Scholar]
- Bertolino E, Reimund B, Wildt-Perinic D, Clerc RG. A novel homeobox protein which recognizes a TGT core and functionally interferes with a retinoid-responsive motif. J Biol Chem. 1995;270:31178–31188. doi: 10.1074/jbc.270.52.31178. [DOI] [PubMed] [Google Scholar]
- Bürglin TR. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 1997;25:4173–4180. doi: 10.1093/nar/25.21.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürglin TR. The PBC domain contains a MEINOX domain: coevolution of Hox and TALE homeobox genes? Dev Genes Evol. 1998;208:113–116. doi: 10.1007/s004270050161. [DOI] [PubMed] [Google Scholar]
- Bürglin TR, Mukherjee K. Comprehensive analysis of animal TALE homeobox genes: new conserved motifs and cases of accelerated evolution. J Mol Evol. 2007;65:137–153. doi: 10.1007/s00239-006-0023-0. [DOI] [PubMed] [Google Scholar]
- Anderson DM, Arredondo J, Hahn K, Valente G, Martin JF, Wilson-Rawls J, Rawls A. Mohawk is a novel homeobox gene expressed in the developing mouse embryo. Dev Dynam. 2006;235:792–801. doi: 10.1002/dvdy.20671. [DOI] [PubMed] [Google Scholar]
- Harada R, Bérubé G, Tamplin OJ, Denis-Larose C, Nepveu A. DNA-binding specificity of the cut repeats from the human cut-like protein. Mol Cell Biol. 1995;15:129–140. doi: 10.1128/mcb.15.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürglin TR, Cassata G. Loss and gain of domains during evolution of cut superclass homeobox genes. Int J Dev Biol. 2002;46:115–123. [PubMed] [Google Scholar]
- Yousef MS, Matthews BW. Structural basis of prospero-DNA interaction: implications for transcription regulation in developing cells. Structure. 2005;13:601–607. doi: 10.1016/j.str.2005.01.023. [DOI] [PubMed] [Google Scholar]
- Nishijima I, Ohtoshi A. Characterization of a novel prospero-related homeobox gene, Prox2. Mol Gen Genom. 2006;275:471–478. doi: 10.1007/s00438-006-0105-0. [DOI] [PubMed] [Google Scholar]
- Bayarsaihan D, Enkhmandakh B, Makeyev A, Greally JM, Leckman JF, Ruddle FH. Homez, a homeobox leucine zipper gene specific to the vertebrate lineage. Proc Natl Acad Sci USA. 2003;100:10358–10363. doi: 10.1073/pnas.1834010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelles L, Van de Putte T, van Grunsven L, Huylebroeck D, Verschueren K. Organization of the mouse Zfhx1b gene encoding the two-handed zinc finger repressor Smad-interacting protein-1. Genomics. 2003;82:460–469. doi: 10.1016/S0888-7543(03)00169-1. [DOI] [PubMed] [Google Scholar]
- Mizutani Y, Kihara A, Igarashi Y. Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem J. 2005;390:263–271. doi: 10.1042/BJ20050291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pewzner-Jung Y, Ben-Dor S, Futerman AH. When do Lasses (longevity assurance genes) become CerS (ceramide synthases)?: Insights into the regulation of ceramide synthesis. J Biol Chem. 2006;281:25001–25005. doi: 10.1074/jbc.R600010200. [DOI] [PubMed] [Google Scholar]
- Rao E, Weiss B, Fukami M, RumpAndreas , Niesler B, Mertz A, Muroya K, Binder G, Kirsch S, Winkelmann M, et al. Pseudoautosomal deletions encompassing a novel homeobox gene cause growth failure in idiopathic short stature and Turner syndrome. Nat Genet. 1997;16:54–63. doi: 10.1038/ng0597-54. [DOI] [PubMed] [Google Scholar]
- Blanco-Arias P, Sargent CA, Affara NA. The human-specific Yp11.2/Xq21.3 homology block encodes a potentially functional testis-specific TGIF-like retroposon. Mamm Genome. 2002;13:463–468. doi: 10.1007/s00335-002-3010-9. [DOI] [PubMed] [Google Scholar]
- NCBI BLAST http://www.ncbi.nlm.nih.gov/BLAST/
- McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JPred http://www.compbio.dundee.ac.uk/Software/JPred/jpred.html
- Ensembl Genome Browser http://www.ensembl.org/
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP: Phylogeny Inference Package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- TreeExplorer http://evolgen.biol.metro-u.ac.jp/TE/TE_man.html
- Liu M, Su M, Lyons GE, Bodmer R. Functional conservation of zinc-finger homeodomain gene zfh1/SIP1 in Drosophila heart development. Dev Genes Evol. 2006;216:683–693. doi: 10.1007/s00427-006-0096-1. [DOI] [PubMed] [Google Scholar]
- Manfroid I, Caubit X, Kerridge S, Fasano L. Three putative murine Teashirt orthologues specify trunk structures in Drosophila in the same way as the Drosophilateashirt gene. Development. 2004;131:1065–1073. doi: 10.1242/dev.00977. [DOI] [PubMed] [Google Scholar]
- Caubit X, Coré N, Boned A, Kerridge S, Djabali M, Fasano L. Vertebrate orthologues of the Drosophila region-specific patterning gene teashirt. Mech Dev. 2000;91:445–448. doi: 10.1016/S0925-4773(99)00318-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Maximum likelihood phylogenetic tree of all human plus selected protostome and cnidarian homeodomains for identification of gene families. Arbitrarily rooted phylogenetic tree of all human plus selected protostome and cnidarian homeodomains constructed using the maximum likelihood (ML) method. Bootstrap values supporting gene family designations are shown. Homeodomain sequences derived from pseudogenes are excluded. This ML tree should be compared with the neighbor-joining (NJ) tree shown in Additional file 2. The dataset used for both ML and NJ analyses includes all human homeodomains, most Drosophila melanogaster homeodomains, plus selected additional homeodomains from other protostomes or cnidarians when the gene family is divergent or absent in Drosophila. Divergent Drosophila genes that do not group with human genes were identified by construction of a preliminary, non-bootstrapped ML and NJ trees, and subsequently removed from the dataset. These include genes lost from human, as well genes known to have undergone unusually rapid evolution in Drosophila. For the Hox3 family the rapidly evolving Drosophila genes bcd, zen and zen2 were replaced with Sm Hox3b, and for the Nk4 family the rapid evolving Drosophila gene tin was replaced with Pd NK4. In addition, six genes from other protostome or cnidarian genomes were added to represent gene families known to be missing from Drosophila (Pdx family: Ps Xlox; Alx family: Nv CART1; Dmbx family: Hv manacle; Pou1 family: Nv POU1; Hnf1 family: Nv HNF; Pknox family: Am Prep). Species abbreviations: Am, Apis mellifera (honeybee); Dm, Drosophila melanogaster (fruitfly); Hv, Hydra vulgaris (hydrozoan); Nv, Nematostella vectensis (starlet sea anemone); Pd, Platynereis dumerilii (annelid worm); Ps, Phascolion strombus (sipunculan worm); Sm, Strigamia maritima (centipede). ML performed more poorly than NJ in recovering several well known gene families, notably Hox4, Hox5, Nk4 and Alx. In contrast, ML did recover PROP1 and CG32532 as a true gene family; NJ did not. The invertebrate gene does not always lie as a strict outgroup to all human genes in a family; this effect is expected when using a short alignment. Instead, distinct grouping of invertebrate and human genes is taken as evidence of ancestry from a single gene. A few ambiguous cases were encountered, notably divergence of Drosophila H2.0 in the proposed Hlx gene family, and resolution within the Pax4/6 gene family, which is recovered as two families in NJ but one in ML. As explained in the text, several human gene families contain 'orphan' genes without invertebrate orthologs; these are Barx, Nanog, Noto, Vax, Ventx, Argfx, Dprx, Dux, Esx, Hesx, Hopx, Isx, Leutx, Mix, Nobox, Rhox, Sebox, Tprx, Hdx, Pou5, Hmbox, Satb, Adnp and Zhx/Homez. Zeb and Mkx would be placed in this category based on our ML and NJ trees, although other data suggest that Drosophila zfh1 and CG11617 respectively may be the protostome orthologs [73,94]. Tshz is only an apparent orphan family; the clear Drosophila ortholog simply lacks the homeobox [95,96]. Phylogenetic analysis is just one source of evidence for allocation of genes to gene families and identification of boundaries between gene families; complementary criteria used are synteny between species and paralogy within the human genome. Our ML and NJ trees should not be used to allocate gene families to gene classes, because other diagnostic characters such as insertions within the homeodomain, key amino acid residues, and several motifs outside of the homeodomain are excluded from the analysis. Indeed, artefactual mixing of the TALE and SINE classes occurs in both ML and NJ trees.
Neighbor-joining phylogenetic tree of all human plus selected protostome and cnidarian homeodomains for identification of gene families. Arbitrarily rooted phylogenetic tree of all human plus selected protostome and cnidarian homeodomains constructed using the neighbor-joining (NJ) method. Bootstrap values supporting gene family designations are shown. Homeodomain sequences derived from pseudogenes are excluded. Comparison of NJ and ML trees, and description of the dataset used, is given in the legend to Additional file 1. Several artefactual mixing of classes occurs in this NJ tree, notably splitting of the CUT class, mixing of the TALE and SINE classes and aberrant placement of HOPX.
Neighbor-joining phylogenetic tree of human ANTP-class homeodomains, for comparison to maximum likelihood tree. Arbitrarily rooted phylogenetic tree of human ANTP-class homeodomains constructed using the neighbor-joining method. Bootstrap values supporting internal nodes with over 70% are shown. Homeodomain sequences derived from pseudogenes are excluded. The proposed division between the HOXL and NKL subclasses is indicated. The position of EN1 and EN2 is unstable; this tree places them close to the base of the HOXL/NKL divergence, whereas maximum likelihood analysis of the same dataset places them firmly in the NKL subclass (Figure 1). Interrelationships of genes in the Nk2.2 and Nk4 families are also unstable (in this tree and Figure 1 respectively); in these cases synteny within and between genomes clearly resolves gene families. Detailed relationships between different gene families should not be inferred from this tree.
Neighbor-joining phylogenetic tree of human PRD-class homeodomains, for comparison to maximum likelihood tree. Arbitrarily rooted phylogenetic tree of human PRD-class homeodomains constructed using the neighbor-joining method. Bootstrap values supporting internal nodes with over 70% are shown. Homeodomain sequences derived from pseudogenes are excluded, as are the partial homeodomains of PAX2, PAX5 and PAX8, and the HOPX homeodomain because its extremely divergent sequence destabilizes the overall tree topology. Roman numeral suffixes are used to distinguish multiple homeodomains encoded by a single Dux-family gene. Detailed relationships between different gene families should not be inferred from this tree.
Neighbor-joining phylogenetic tree of human homeodomains excluding ANTP and PRD classes, for comparison to maximum likelihood tree. Arbitrarily rooted phylogenetic tree of human homeodomains excluding the ANTP and PRD classes constructed using the neighbor-joining method. Bootstrap values supporting internal nodes with over 70% are shown. Homeodomain sequences derived from pseudogenes are excluded. Roman numeral suffixes are used to distinguish multiple homeodomains encoded by a single gene. Classes and/or families are color coded as shown in the key. The LIM and ZF classes are not recovered as two distinct monophyletic groups, a result also found by maximum likelihood analysis (Figure 3). The multiple homeodomains of Zfhx-family proteins and Zhx/Homez-family proteins are also dispersed in the tree, presumably artefactually. Monophyly of the CUT class is not recovered in this tree, but is by maximum likelihood analysis (Figure 3). Detailed relationships between different gene families should not be inferred from this tree.
Multiple sequence alignment of all human plus selected protostome and cnidarian homeodomains. The consensus homeodomain sequence (shown several times for reference) was derived from a compilation of 247 human homeodomain sequences. The three horizontal lines indicate the positions of the three alpha-helices. The numbering scheme refers to amino acid position in the canonical 60-amino-acid homeodomain; insertions relative to this sequence are shown when present. Black shaded resides are invariant between all human homeodomains within each class (or family in the case of the ZF homeodomains). Sequence accession numbers are shown. For each gene family designation, maximum likelihood and neighbor-joining bootstrap support values are indicated (see Additional files 1 and 2). These values are not shown if the gene family does not form a monophyletic group in phylogenetic analyses (in which case n/a is written) or if an invertebrate homolog could not be found.
Phylogenetic input file. All human and invertebrate homeodomains used in phylogenetic analyses are shown, after alignment and removal of insertions to give a uniform 60-amino-acid alignment.