Figure 1.
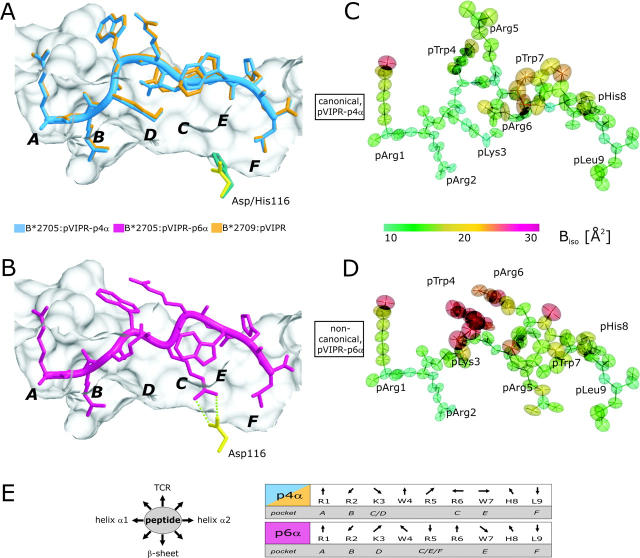
pVIPR conformations, atomic displacement ellipsoids, and B factors. (A) Superimposition of the canonical pVIPR conformations (p4α) found in B*2705 (blue) and B*2709 (gold). (B) The noncanonical pVIPR conformation (p6α, pink) observed only in B*2705. The peptides are viewed from the side of the α2 helix together with a molecular surface covering the floor and back of the binding groove. The subtype-specific residue 116 is indicated also (Asp116, yellow; His116, turquoise); the bidentate salt bridge to Asp116 is drawn with green dotted lines in B. The binding pockets A–F are shown in bold letters. Atomic displacement ellipsoids for pVIPR-p4α and -p6α in C and D are colored according to the equivalent isotropic temperature factors B (Å2) (see color bar). (E, left) Schematic description of side chain orientation when looking from the NH2 to the COOH terminus of pVIPR. Bottom of peptide binding groove indicated by “β-sheet” and side for T cell recognition by “TCR”. (E, right) The orientation of the peptide side chains in the p4α and p6α conformations as in E (left). The respective binding pockets (A–F) are indicated as well. It is clear from this representation and Fig. 1 (A and B) that the two pVIPR conformations show major differences only from pLys3 to pTrp7.