Abstract
Enhancers regulate lineage choice and the developmental timing of antigen receptor gene rearrangements. The transcription factor NF-κB has been implicated as a key component of the recombination and transcription activation potential of the immunoglobulin κ chain gene intronic enhancer. Here, I discuss the implications of the new observation that an NF-κB binding site–mutated enhancer in the correct biological context does not appear to affect κgene expression.
Programmed rearrangement of antigen receptor gene loci is a hallmark of B and T lymphocyte development. Of the two chains that make up the receptors, the Ig heavy chain (IgH) and the TCRβ chain loci are the first to rearrange in the B and T lineages, respectively. The production of functional genes requires two recombination events that juxtapose V, D, and J gene segments. This is followed by Ig light chain (IgL) or TCRα chain gene recombination, which involves a single step that joins a V and J gene segment. The error-prone nature of the V(D)J recombinase results in only a small proportion of pre–B or pro–T cells generating a functional IgH or TCRβ gene that can express protein. The minority of cells that express IgH or TCRβ proteins are selected to proliferate and differentiate further via signals from pre–B or pre–T cell receptors. Single step recombination at the second antigen receptor locus (IgL or TCRα) and the ability to undergo sequential rearrangements makes the chances of producing a functional gene at these loci much higher. Thus, the order of gene rearrangements ensures that most cells that express IgH or TCRβ will successfully recombine IgL or TCRα genes and express a complete antigen receptor.
Developmental stage-specific recombination of antigen receptor loci is regulated by cis-regulatory enhancer sequences that alter accessibility of the recombinase machinery to the gene segments. The function of enhancers is mediated by DNA binding proteins that recruit to the enhancer, via protein–protein interactions, a multisubunit complex that is a functional enhancer.
Transcriptional Regulation of the κ Locus.
NF-κB binds to the DNA sequence known as the κB site within the enhancer (iEκ) located in the Jκ-Cκ intron of the Ig κ light chain gene (Fig. 1 and reference 1). The presence of nuclear NF-κB DNA binding activity in Ig-expressing B lymphocytes, but not at earlier stages of B cell differentiation, suggested that it might play a key role in κ light chain gene expression. This idea was reinforced by observations that NF-κB induction in pre–B cell lines coordinately activates κ gene transcription and Vκ to Jκ recombination (2, 3). Conversely, both processes are adversely affected when NF-κB activation is blocked (4).
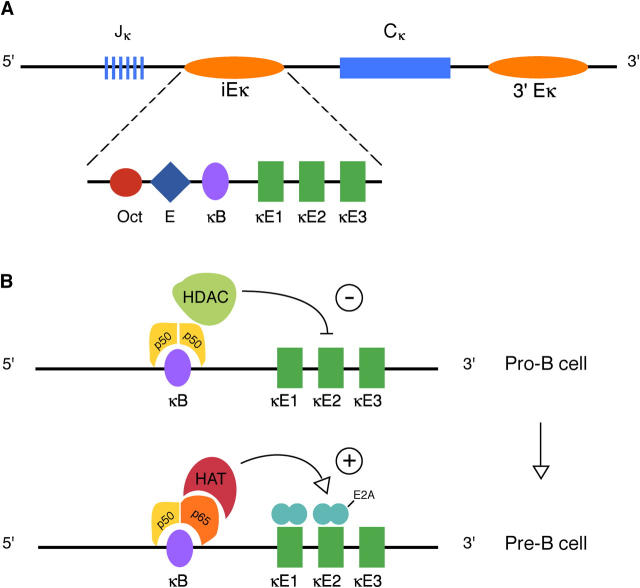
Figure 1.
Regulation of the κ locus. (A) Organization of the κ light chain locus showing the Jκ gene segments, intronic enhancer (iEκ), the constant region exons, and the 3′ enhancer (3′Eκ). The iEκ contains binding sites for NF-κB (κB), E2A (κE1, κE2), and other transcription factors. (B) Hypothetical model of NF-κB–mediated repression and de-repression of iEκ. Binding of p50 homodimers to the κB site at the pro–B stage results in recruitment of HDACs and inhibition of E2A-driven transcription (top). Replacement of p50 homodimers by p50-p65 heterodimers at the pro–B to pre–B cell transition recruitsHATs which counteract the repression and allow gene activation by E2A proteins (bottom).
In addition to κB, the iEκ contains binding sites for several other transcription factors (Fig. 1 A). Among these, κE1-κE3 share a CAXXTGG sequence motif that binds proteins of the basic helix-loop-helix (bHLH) family. κE1 and κE2 bind E2A proteins, whereas κE3 binds TFE3 and the closely related protein USF, which are leucine zipper containing bHLH proteins. Functional analyses of motifs within iEκ performed using isolated enhancers to activate reporter genes in transfection assays have shown that B cell–specific transcriptional activity of the enhancer depends substantially on the κB element (5). Moreover, iEκ is inactive in pre–B cell lines except when the cells are treated with LPS, an agent that activates NF-κB DNA binding. Mutations of κE1-κE3 have variable and weaker effects on transcription compared to mutation of the κB site. The κB site is also essential for the activation of recombination by iEκ (6) and as an element that promotes B cell–specific CpG demethylation of transfected genes (an epigenetic modification that correlates with increased transcriptional activity) (7). Cumulatively, these observations strengthened the idea of NF-κB as the master and commander of κ gene expression via the κB motif in iEκ.
The past two decades have witnessed enormous growth in the number of biological roles ascribed to NF-κB. First described as a κ gene–activating protein in lymphocytes, it is now known to function in a wide variety of physiological and pathologic processes in diverse cell types (8, 9). Perhaps as a result, the role of the κB site in the κ enhancer was somewhat neglected. In this issue, Inlay et al. perform the definitive experiment to determine the role of the κB motif in its normal context (10). They used gene targeting to knock in mutated iEks at the endogenous locus and then used the mutant embryonic stem cells to complement Rag-deficient blastocysts. The status of κ gene rearrangements was compared on the wild-type and mutated alleles in mature heterozygote B cells. Surprisingly, the allele bearing the κB site mutation recombined at virtually wild-type levels; in contrast, alleles bearing κE1 or κE2 mutations rearranged less efficiently. Double mutation of the κE1 and κE2 sites markedly reduced rearrangement to levels seen after deletion of iEκ (11). The authors conclude that E2A proteins that bind κE1 and κE2 are essential for efficient κ gene rearrangements. In contrast, the κB site and NF-κB appears to be unimportant for κ gene recombination.
A Role for NF-κB in Epigenetic Regulation of Rearrangement?
NF-κB–dependent regulation of the κ locus remains a viable proposition, in my opinion, because it provides a likely mechanism for the developmental timing of κ gene rearrangements. Though low levels have been noted in pro–B cells, the majority of the κ rearrangements occur in the pre–B compartment after IgH-expressing cells are selected by the pre–B cell receptor (pre-BCR). This bias toward late rearrangements of κ genes is not easily explained by an E2A-only mechanism because these bHLH proteins are expressed in pro–B cells where they activate IgH rearrangements via binding sites in the J H-Cμ intron enhancer (Eμ) (12). If E2A proteins are sufficient to activate iEκ and are present and functional from the earliest stages of B cell development, what prevents κ genes from recombining before the pre–B cell stage? It is possible that E2A functions on the enhancer may be regulated differentially in pro–B cells and pre–B cells. For example, increased levels of expression or posttranslational modifications may increase the function of E2A on iEκ in pre–B cells compared with pro–B cells. However, there is little evidence for such changes between pro–B and pre–B cells. Instead, there is ample evidence for NF-κB activation via the pre-BCR during the pro–B to pre–B cell transition (13, 14).
How might NF-κB activation render iEκ more susceptible to E2A-dependent activation at the correct developmental stage? One possibility is that Rel proteins (the subunits that make up the NF-κB dimers) bind to the κB site in pro–B cells (15) to prevent E2A-mediated activation of recombination (Fig. 1 B). The most likely negative regulator is the p50 homodimer. This Rel protein does not contain a classical activation domain and has been shown to recruit histone deacetylases (HDACs) to regulatory sequences to actively repress transcription by epigenetic means (16). Under some circumstances, p65 (also known as RelA) behaves similarly (17), although it is usually associated with gene activation. Pre-BCR–induced NF-κB activation in pre–B cells may tilt the balance toward de-repression of the κ locus by replacing p50 homodimers with Rel proteins that contain transcription activation domains (such as p65 or c-Rel). These Rel proteins can recruit histone acetyl transferases (HATs) (18, 19) to counteract the effects of histone deacetylases located on the gene, resulting in gene activation. The crux of the hypothesis is to view the role of NF-κB at the κ locus as mediating repression or de-repression depending on the stage of B cell differentiation, with the outcome being determined by different sets of Rel proteins. In this model, mutation of the κB site prevents binding of Rel proteins that initiate repression in pro–B cells, as a result of which the timing of κ gene rearrangements may be altered. In the simplest scenario, κ recombination may occur earlier during B cell differentiation. Such NF-κB–independent recombination may be mechanistically analogous to that induced by ectopic E2A expression in nonlymphoid cells (20, 21).
If the κB site prevents E2A-dependent recombination in pro–B cells, the κB mutant allele in the study by Inlay et al. (10) might be expected to recombine at higher levels than the wild-type allele because it would be accessible in both pro–B cells and pre–B cells. However, without direct analysis of the pro–B compartment, it may be difficult to discern whether recombination is increased, given the small numbers of pro–B cells relative to pre–B cells. Alternatively, the comparable levels of recombination on both the κB mutant and wild-type alleles in mature heterozygote B cells (10) suggests that the κE1- plus κE2-driven enhancer may not be effectively activated by the nuclear milieu of the pro–B cell. What could be missing in pro–B cells that later compensates for the loss of the κB site in pre–B cells? One possibility is the 3′k enhancer. Once activated in pre–B cells, it may increase the recombination potential of the “crippled” κB mutant iEκ, in a way serving as a surrogate for the missing NF-κB that would normally be recruited to the locus. In the system developed by Inlay et al., this could be tested by analyzing the effect of the κB mutation in the absence of 3′Eκ (10). Clearly, however, the 3′Eκ cannot rescue a κE1 plus κE2 doubly mutated iEκ. The proposed insufficiency of a κB-mutated enhancer in the absence of additional positive regulatory sequences is consistent with all earlier functional studies of the isolated iEκ.
Further Implications.
Epigenetic repression and de-repression via NF-κB proteins provides plausible explanations for several other aspects of κ gene regulation. First, the κ locus in a minority of pro–B cells may escape p50-dependent epigenetic silencing. Inefficient activation of iEκ via E2A proteins in these cells may result in the low level of κ recombination seen in pro–B cells. Second, de-repression in pre–B cells may be limited by the concentration of HAT-recruiting Rel family members activated by the pre-BCR. It is possible that pre-BCR signaling is not strong enough or does not last long enough to generate enough positive-acting NF-κB. Thus, de-repression might occur only in that subset of cells where the level of activating Rel proteins reaches a threshold required to counteract repression. This may be the basis for variegated κ gene expression in the pre–B compartment that was described recently by Liang et al. (22). It is worth noting that the more uniform expression of κ genes in mature B cells probably reflects transcription activation via the 3′Eκ. Thus, each of the two enhancers might serve different, though overlapping, functions; iEκ is primarily involved in activating Vκ recombination in pre–B cells, though its deficiency can be partially, or fully, compensated depending on the severity of the mutation, whereas the 3′Eκ is primarily involved in activating κ gene transcription. Third, the κB site–dependent epigenetic changes proposed here must be superimposed on mono-allelic DNA methylation of the κ locus (23), a modification that is also associated with HDAC recruitment and gene silencing (24). Thus, multiple levels of epigenetic regulation may control the timed onset of κ gene recombination.
Overall, Inlay et al.'s clean experiment and its unexpected result proves to be more thought provoking than anticipated. Most importantly, it demonstrates that the timing of κ gene expression during B cell development remains an open question, be it mediated by NF-κB or by a different, presently unknown mechanism.
Acknowledgments
I thank Drs. Yehudit Bergman and Mark Schlissel for taking the time to share their expertise in κ gene regulation during preparation of this commentary.
References
- 1.Calame, K., and R. Sen. 2003. Transcription of immunoglobulin genes. Molecular Biology of B Cells. T. Honjo, F. Alt, and M. Neuberger, editors. Elsevier Science Publishing Co. Inc., New York. 79–96 pp.
- 2.Schlissel, M.S., and D. Baltimore. 1989. Activation of immunoglobulin kappa gene rearrangement correlates with induction of germline kappa gene transcription. Cell. 58:1001–1007. [DOI] [PubMed] [Google Scholar]
- 3.Klug, C.A., S.J. Gerety, P.C. Shah, Y.Y. Chen, N.R. Rice, N. Rosenberg, and H. Singh. 1994. The v-abl tyrosine kinase negatively regulates NF-kappa B/Rel factors and blocks kappa gene transcription in pre-B lymphocytes. Genes Dev. 8:678–687. [DOI] [PubMed] [Google Scholar]
- 4.Scherer, D.C., J.A. Brockman, H.H. Bendall, G.M. Zhang, D.W. Ballard, and E.M. Oltz. 1996. Corepression of RelA and c-rel inhibits immunoglobulin kappa gene transcription and rearrangement in precursor B lymphocytes. Immunity. 5:563–574. [DOI] [PubMed] [Google Scholar]
- 5.Lenardo, M., J.W. Pierce, and D. Baltimore. 1987. Protein-binding sites in Ig gene enhancers determine transcriptional activity and inducibility. Science. 236:1573–1577. [DOI] [PubMed] [Google Scholar]
- 6.Demengeot, J., E.M. Oltz, and F.W. Alt. 1995. Promotion of V(D)J recombinational accessibility by the intronic E kappa element: role of the kappa B motif. Int. Immunol. 7:1995–2003. [DOI] [PubMed] [Google Scholar]
- 7.Kirillov, A., B. Kistler, R. Mostoslavsky, H. Cedar, T. Wirth, and Y. Bergman. 1996. A role for nuclear NF-kappaB in B-cell-specific demethylation of the Igkappa locus. Nat. Genet. 13:435–441. [DOI] [PubMed] [Google Scholar]
- 8.Amit, S., and Y. Ben-Neriah. 2003. NF-kappaB activation in cancer: a challenge for ubiquitination- and proteasome-based therapeutic approach. Semin. Cancer Biol. 13:15–28. [DOI] [PubMed] [Google Scholar]
- 9.Smahi, A., G. Courtois, S.H. Rabia, R. Doffinger, C. Bodemer, A. Munnich, J.L. Casanova, and A. Israel. 2002. The NF-kappaB signalling pathway in human diseases: from incontinentia pigmenti to ectodermal dysplasias and immune-deficiency syndromes. Hum. Mol. Genet. 11:2371–2375. [DOI] [PubMed] [Google Scholar]
- 10.Inlay, M.A., H. Tian, T. Lin, and Y. Xu. Important roles for E protein binding sites within the immunoglobulin κ chain intronic enhancer in activating Vκ Jκ rearrangement. J. Exp. Med. 200:1205–1211. [DOI] [PMC free article] [PubMed]
- 11.Xu, Y., L. Davidson, F.W. Alt, and D. Baltimore. 1996. Deletion of the Ig kappa light chain intronic enhancer/matrix attachment region impairs but does not abolish V kappa J kappa rearrangement. Immunity. 4:377–385. [DOI] [PubMed] [Google Scholar]
- 12.Kee, B.L., M.W. Quong, and C. Murre. 2000. E2A proteins: essential regulators at multiple stages of B-cell development. Immunol. Rev. 175:138–149. [PubMed] [Google Scholar]
- 13.Saijo, K., C. Schmedt, I.H. Su, H. Karasuyama, C.A. Lowell, M. Reth, T. Adachi, A. Patke, A. Santana, and A. Tarakhovsky. 2003. Essential role of Src-family protein tyrosine kinases in NF-kappaB activation during B cell development. Nat. Immunol. 4:274–279. [DOI] [PubMed] [Google Scholar]
- 14.Tretter, T., A.E. Ross, D.I. Dordai, and S. Desiderio. 2003. Mimicry of pre–B cell receptor signaling by activation of the tyrosine kinase Blk. J. Exp. Med. 198:1863–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaffer, A.L., A. Peng, and M.S. Schlissel. 1997. In vivo occupancy of the kappa light chain enhancers in primary pro- and pre-B cells: a model for kappa locus activation. Immunity. 6:131–143. [DOI] [PubMed] [Google Scholar]
- 16.Zhong, H., M.J. May, E. Jimi, and S. Ghosh. 2002. The phosphorylation status of nuclear NF-kappa B determines its association with CBP/p300 or HDAC-1. Mol. Cell. 9:625–636. [DOI] [PubMed] [Google Scholar]
- 17.Ashburner, B.P., S.D. Westerheide, and A.S. Baldwin Jr. 2001. The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol. Cell. Biol. 21:7065–7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong, H., R.E. Voll, and S. Ghosh. 1998. Phosphorylation of NF-kappa B p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol. Cell. 1:661–671. [DOI] [PubMed] [Google Scholar]
- 19.Sheppard, K.A., D.W. Rose, Z.K. Haque, R. Kurokawa, E. McInerney, S. Westin, D. Thanos, M.G. Rosenfeld, C.K. Glass, and T. Collins. 1999. Transcriptional activation by NF-kappaB requires multiple coactivators. Mol. Cell. Biol. 19:6367–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romanow, W.J., A.W. Langerak, P. Goebel, I.L. Wolvers-Tettero, J.J. van Dongen, A.J. Feeney, and C. Murre. 2000. E2A and EBF act in synergy with the V(D)J recombinase to generate a diverse immunoglobulin repertoire in nonlymphoid cells. Mol. Cell. 5:343–353. [DOI] [PubMed] [Google Scholar]
- 21.Goebel, P., N. Janney, J.R. Valenzuela, W.J. Romanow, C. Murre, and A.J. Feeney. 2001. Localized gene-specific induction of accessibility to V(D)J recombination induced by E2A and early B cell factor in nonlymphoid cells. J. Exp. Med. 194:645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang, H.E., L.Y. Hsu, D. Cado, and M.S. Schlissel. 2004. Variegated transcriptional activation of the immunoglobulin kappa locus in pre-b cells contributes to the allelic exclusion of light-chain expression. Cell. 118:19–29. [DOI] [PubMed] [Google Scholar]
- 23.Mostoslavsky, R., N. Singh, A. Kirillov, R. Pelanda, H. Cedar, A. Chess, and Y. Bergman. 1998. Kappa chain monoallelic demethylation and the establishment of allelic exclusion. Genes Dev. 12:1801–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bird, A.P., and A.P. Wolffe. 1999. Methylation-induced repression–belts, braces, and chromatin. Cell. 99:451–454. [DOI] [PubMed] [Google Scholar]