Figure 2.
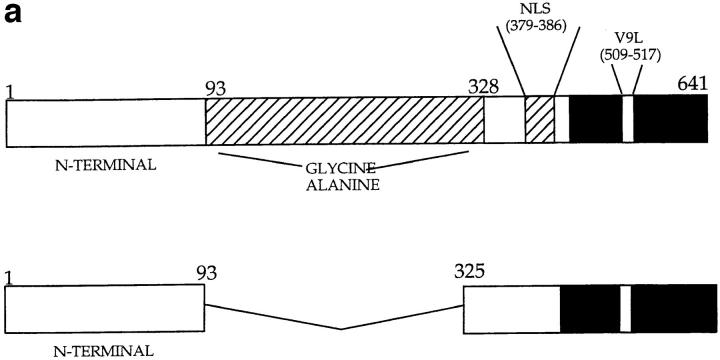
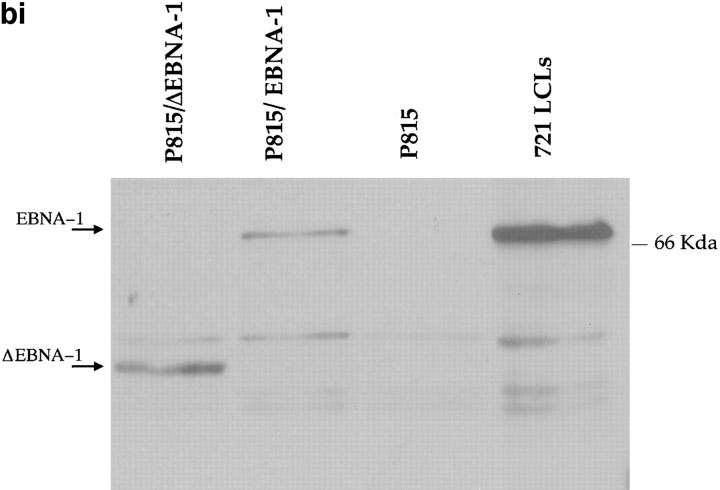
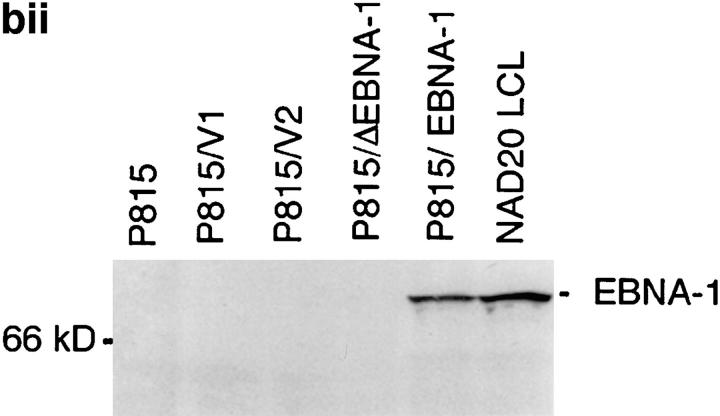
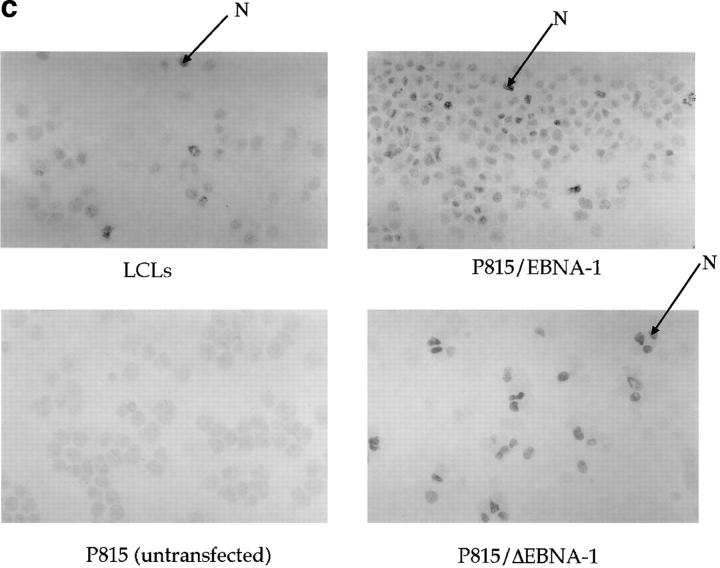
(a) Constructs used to transfect P815 cells. (Top) Full-length EBNA-1 of the B-95-8 sequence (641 amino acids total). Hatched region (93–328) shows the Gly-Ala–rich region. NLS, nuclear localisation sequence as described in reference 16. The position of the V9L epitope is 509–517 (indicated as V9L). (Bottom) ΔEBNA-1: residues 93–325 have been deleted, but the rest of EBNA-1 is unaffected. (b, i) Western Blot analysis of transfected cells using the Rbt-EBNA-1 antibody (11). Cells are as labeled. A 72-kD band is detected in P815/EBNA-1 cells; based on reactivity with the antiserum and the size of EBNA-1 detected in the 721 LCL line (lane 4), this is EBNA-1. A 43-kD band is detected in P815 cells transfected with EBNA-1 lacking the Gly-Ala repeat. (b, ii) Western Blot using the P-107 serum (12) that reacts against the Gly-Ala repeat only. Lanes 2 and 3 indicate P815 cells transfected with vectors alone (i.e., no EBNA-1). Lane 6, NAD20 cells are EBV-transformed LCLs. (c) Cell staining with Rbt-EBNA-1 serum (11) reactive against EBNA-1. Nuclear staining is indicated with N and arrow. Cells are as indicated in legend and text.