Figure 4.
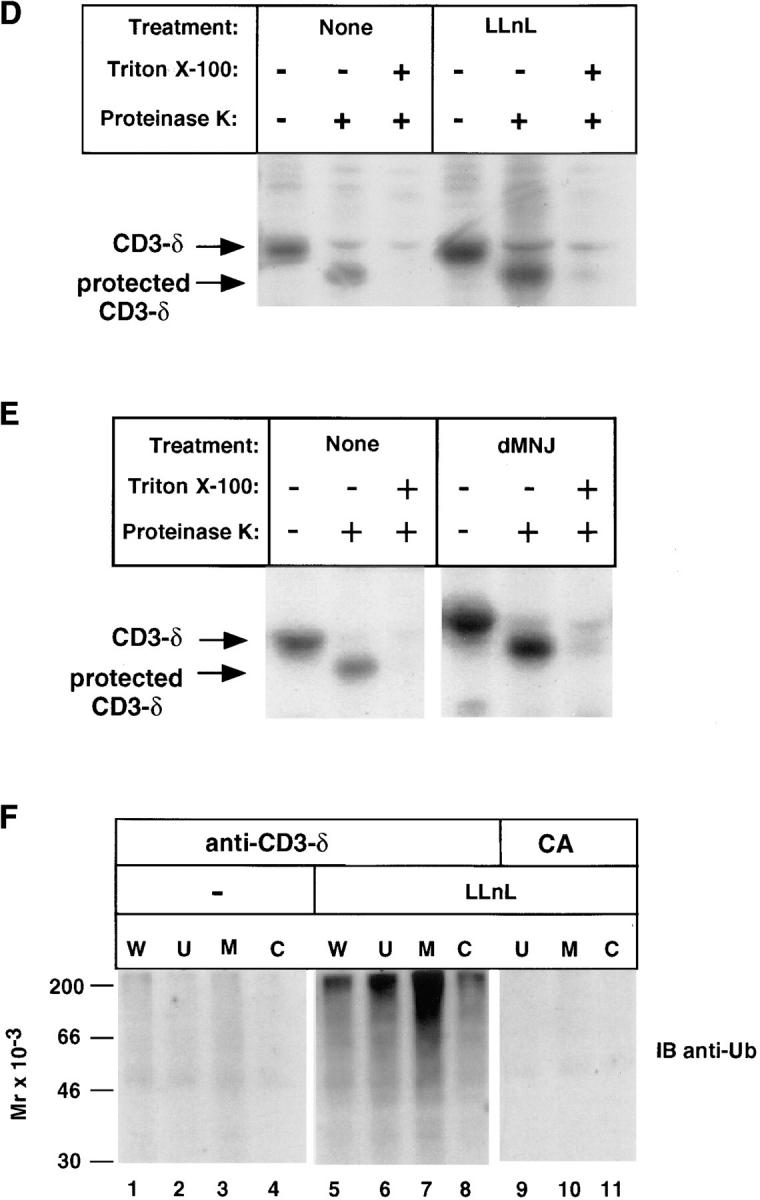
Subcellular localization of CD3-δ. (A) 21.2.2 cells were labeled with [35S]methionine for 20 min followed by a 2.5-h chase. In lanes 1–4, cells were lysed in Triton X-100 lysis buffer directly. In lanes 5–10, cells from the 2.5-h chase were broken open by mechanical shearing without detergent, as described in Materials and Methods, followed by removal of unbroken cells and nuclei (U) and separation of cytosolic (C) and membrane (M) fractions. Immunoprecipitation was with anti–CD3-δ. The amount of material used in the whole cell samples (lanes 1–4) was 1.5 × 107 cells/lane; 3 × 107 cells were subject to fractionation (lanes 5–10). (B) CD3-δ translated in rabbit reticulocytes in vitro and labeled using [35S]methionine was added to cytosol prepared as in A and immunoprecipitated with either CA or anti–CD3-δ. (C) 21.2.2 cells were labeled for 20 min and chased either in the absence or presence of LLnL for 3 h. Membrane (M) and cytosol (C) fractions were prepared as in A. Only half of the residual unbroken cells and nuclei (U) were analyzed in this experiment. Lysates were subject to sequential immunoprecipitation with 2C11(top) followed by anti–CD3-δ (bottom). In pulse-chase experiments CD3-ε labels poorly and is not well-visualized (33). (D) 21.2.2 cells were labeled for 20 min with [35S]methionine, then either treated or not with LLnL for 1 h at 4°C before a 3-h chase in the presence or absence of LLnL. Cells were then broken as in A. After removal of nuclei and unbroken cells, supernatants were divided and treated on ice either without further treatment, with the addition of proteinase K, or first treated with Triton X-100 (0.5%) followed by the addition of proteinase K as indicated. After 1 h at 4°C, PMSF was added to inactivate proteinase K, and Triton X-100 (0.5%) was then added to samples that had not been pretreated with this detergent. All samples were immunoprecipitated with 2C11. The positions of full length CD3-δ and of CD3-δ with a cleaved cytosolic tail are indicated. (E) 21.2.2 cells were subject to pulse-chase metabolic labeling as in C in the presence or absence of dMNJ. Cells were broken, and after removal of unbroken cells and nuclei supernatants were subject to proteinase K digestion and immunoprecipitation with 2C11 as in D. (F) 21.2.2 cells were incubated for 3 h either with or without LLnL. 1.5 × 107 cells were lysed directly in Triton X-100 lysis buffer (W), while the rest (5 × 107) were fractionated as in A followed by immunoprecipitation with anti–CD3-δ. Fractionated LLnL-treated samples were also immunoprecipitated with CA (lanes 9–11). Immunoblotting was with anti-Ub.

