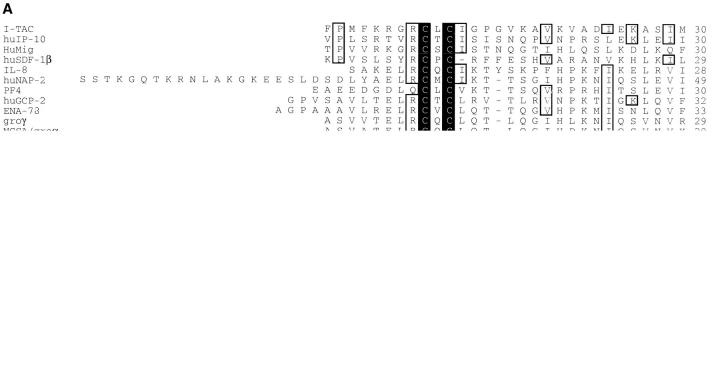
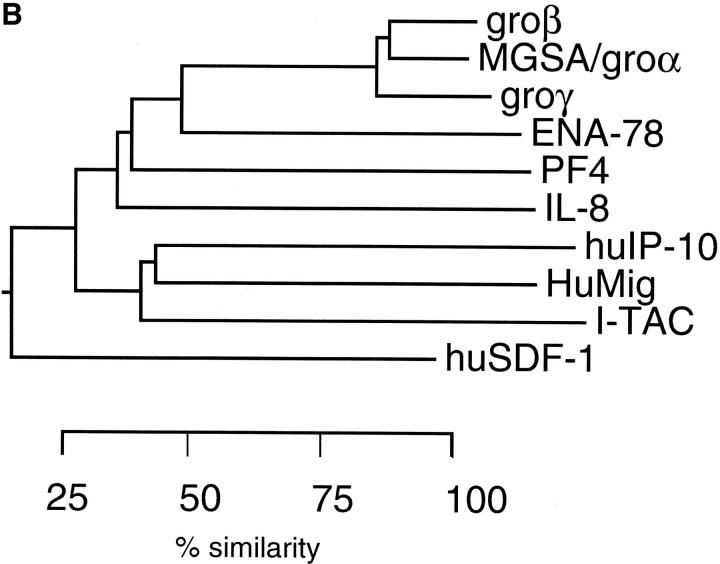
Figure 2.
Sequence alignment and phylogenetic relationship of I-TAC with other known CXC chemokines. (A) Amino acid alignment of I-TAC with CXC chemokines. The alignment was generated with the program Megalign (DNA Star, Madison, WI) using the Clustal method and then manually aligned to obtain the maximum amino acid similarity. Amino acid residues conserved in all sequences are boxed and shaded. Unshaded boxed regions indicate amino acid residues conserved between I-TAC and at least two other CXC chemokines. (B) Phylogenetic tree showing the relationships between I-TAC and known CXC chemokines. The alignment above was used to generate a phylogenetic tree by the Megalign program and the extent of similarity between sequences is indicated below the phylogenetic tree.