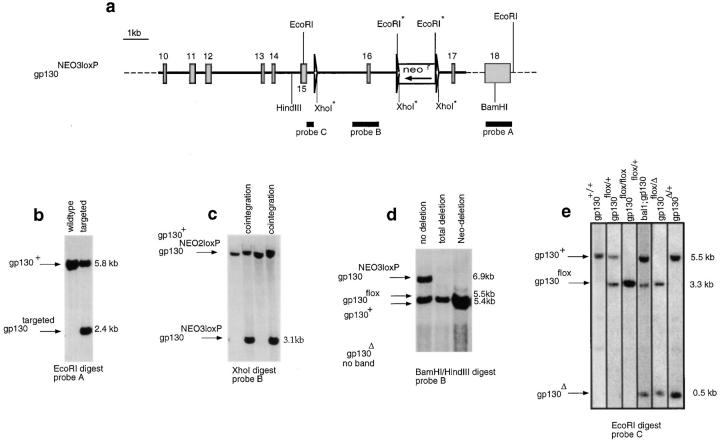
Figure 1.
Gene targeting. (a) Gene targeting vector (bold line) homologously recombined into the gp130 locus (dashed line), thereby generating the targeted allele gp130NEO3loxP. Exon sequences (rectangles); loxP sites (triangles). The transcriptional orientation of the neomycin resistance gene is shown. Restriction sites used for Southern analysis are given; those marked with an asterisk are not present in the wild-type locus but are introduced via the targeting vector. The probes used for Southern analysis are shown (black). In the gp130NEO2loxP allele, the loxP site upstream of exon 16 is missing due to lack of cointegration. (b) Southern analysis to identify homologous recombinant clones. The size and position of the bands representing the gp130+ (wild-type) and gp130targeted (gp130NEO2loxP or gp130NEO3loxP) alleles are given. (c) Southern analysis to identify clones harboring a cointegrated upstream loxP site. The position and size of the bands representing the gp130NEO2loxP and gp130+ alleles or the gp130NEO3loxP allele are given. (d) Southern analysis to identify clones which have deleted the neomycin resistance gene but not exon 16 after transient expression of Cre-recombinase. The position and size of the bands representing the gp130+, gp130flox, and gp130NEO3loxP alleles are given. (e) Southern blot prepared with mouse tail DNA in order to genotype the animals. The position and size of the bands representing the gp130+, gp130flox, and gp130Δ alleles are shown. Lane 4: bal1, Cre-transgenic (reference 31).