Figure 5.
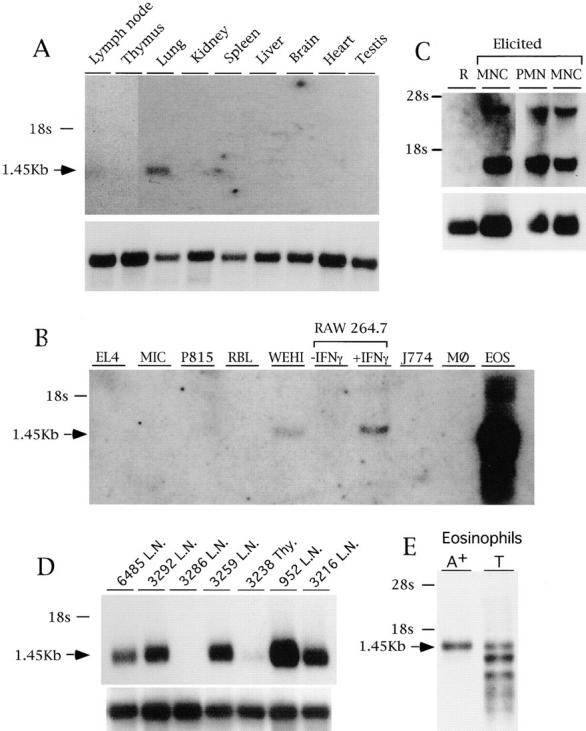
Expression of the mBLTR mRNA. (A) Northern blot analysis of 1.5 μg poly(A)+ RNA isolated from the indicated organs of a healthy FVB mouse. (B) Northern blot analysis of 10 μg total RNA isolated from the EL4 T cell line, MIC B cell line, P815 mastocytoma line, RBL basophilic line, WEHI macrophage line, RAW 264.7 macrophage cell line untreated or treated for 18 h with 200 U/ml IFN-γ, J774 macrophage line, bone marrow derived macrophages (MØ), and eosinophils purified from IL-5 transgenic mice. (C) Northern blot analysis of 10 μg total RNA isolated from leukocytes recovered from peritoneal lavage before (R, resting) or after (elicited) intraperitoneal casein injection and percoll density gradient purification. M, macrophage fraction (76% macrophages, and 24% neutrophils); N, neutrophil fraction (95% neutrophils, 5% macrophages). (D) Northern blot analysis of 1.5 μg poly(A)+ RNA isolated from Thy1 positive fresh lymphomas obtained from lymph nodes (L.N.) or thymus (Thy). Each lane contains RNA isolated from a lymphoma that spontaneously arose in a different bigenic mouse containing a c-myc transgene and p53 null alleles numbered and described in Elson et al. (35). (E) Northern blot analysis of 2 μg of poly(A)+ RNA and 10 μg of total RNA before poly A selection (T) isolated from eosinophils purified from IL-5 transgenic mice. Blots were sequentially probed with the mBLTR (p65b) cDNA (top of each panel) and rpL32 or 28s as a control for RNA loading (bottom of panel).
