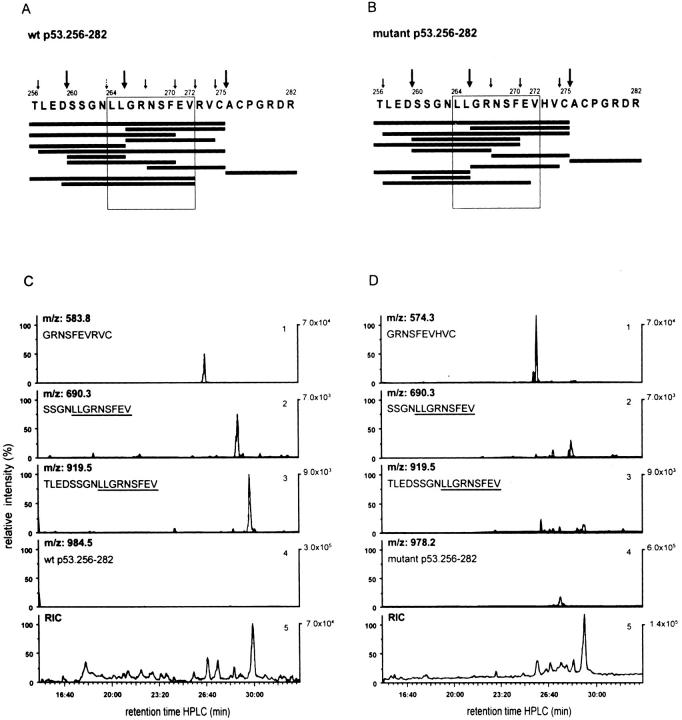
Figure 5.
The p53 hotspot mutation at residue 273 (R to H) abrogates the proteasomal cleavage site between p53 residues 272 and 273. Bulk peptide products derived after 24 h from 20S proteasome–mediated degradation of the synthetic WT (A and C) and mutant (273 R to H) (B and D) 27-mer polypeptides p53.256–282 covering the A*0201-restricted CTL epitope p53.264–272 (LLGRNSFEV) were separated by RP-HPLC and analyzed online by MS. Abundant peptide products were sequenced by MS/MS. Cleavage products with signal intensities of at least threefold above background and identified by mass (MS) and sequence (MS/MS) are shown in descending order according to their signal intensities (A and B, black bars). The amino acid sequence of WT (A) and mutant (B) peptide substrates with abundant (large arrows) and nonabundant (small arrows) cleavage sites is also presented. The small broken arrow (A) represents the theoretical NH2-terminal cleavage site of the nonameric CTL epitope 264–272. The quantitative comparison of some of the relevant WT (C) and mutant (D) cleavage products is shown by their elution profiles and relative signal intensities as measured by the ion current of double protonated peptide ions. As demonstrated in C and D, peptide 266–275 (GRNSFEV-R/H-VC) represents a dominant product that interferes with the formation of the 264–272 CTL epitope (panel 1). The WT p53 peptides 260–272 (SSGNLLGRNSFEV) and 256–272 (TLEDSSGNLLGRNSFEV) use the COOH-terminal cleavage site of the minimal CTL epitope 264–272 between WT residues 272 and 273 (panels 2 and 3). Panel 4 shows the uncleaved WT and mutant 27-mer substrate peptides left after proteasomal degradation and used for adjusting the scale (percentage of relative intensity). Panel 5 gives the total ion current of the bulk proteasomal degradation products.