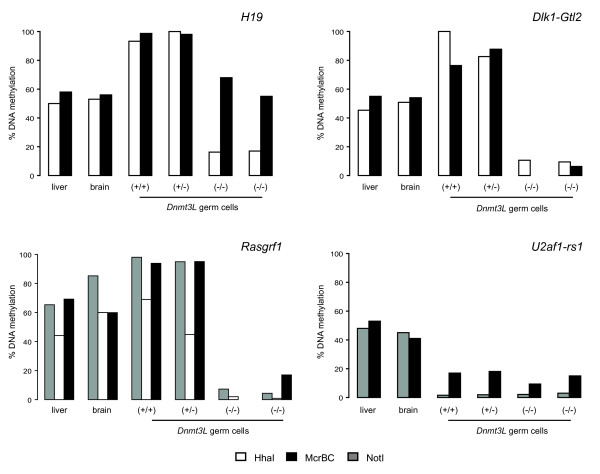
Figure 5.
Abnormal methylation of paternally imprinted genes in Dnmt3L-/- germ cells. DNA extracted from the purified populations of Dnmt3L germ cells was analyzed via a quantitative restriction enzyme assay, quantitative analysis of methylation PCR or qAMP [44]. Analysis of established DMR regions of three known paternally methylated imprinted genes, H19 (top left), Dlk1-Gtl2 (top right) and Rasgrf1 (bottom left), and one known maternally methylated imprinted gene, U2af1-rs1 (bottom right). At least one site of H19, Dlk1-Gtl2 and Rasgrf1 is affected in the two Dnmt3L-/- samples analyzed, whereas U2af1-rs1 methylation remains unchanged. Three different enzymes were used in qAMP: HhaI (white), McrBC (black) and NotI (grey).