FIGURE 2.
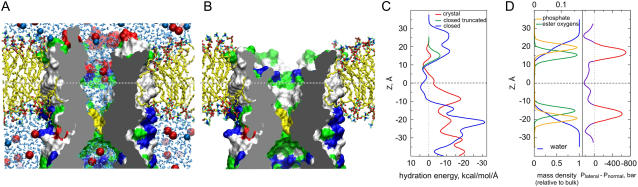
All-atom MD simulation of the modeled resting state and the distribution of polar residues at the protein-lipid boundary. (A) Complete resting state model in the bilayer illustrated by a cross-sectional view captured in the middle of the unrestrained 8 ns simulation (sim2, see also Fig. 3). Shown as solvent-accessible surfaces, polar (green), basic (blue), and acidic (red) residues are exposed to the lipid headgroup regions, whereas nonpolar residues face hydrocarbon. Although the upper hydrophobic chamber is hydrated, neither water (cyan sticks), nor ions (Cl− red or K+ blue, spheres) penetrate the dewetted hydrophobic constriction (yellow). (B) The same structure with the N-terminal domain (26 amino acids) removed. Nicks in the channel wall protrude down to the middle of the fatty acid chains and the absence of lipid-facing polar atoms suggests improper anchoring at the periplasmic rim of the channel. Y27 has spontaneously changed its position during the simulation from pore-facing as in the crystal structure, to one that is more peripheral and solvent-exposed. (C) Hydration energy profile for the lipid-exposed surface reveals a balanced distribution of polar groups in the complete resting state model (blue line) with a hydrophobic region near the midplane of the membrane (shown by horizontal dashed line) that is flanked by polar regions at ±16 Å representing the cytoplasmic and periplasmic rims. The model lacking the N-terminus (green line) shows a very small polar region at the periplasmic rim. Positioning the crystal structure (not shown) in the same way produces a maximum of polarity at −8 Å with a highly imbalanced overall distribution along the Z axis (red line). (D) The distribution of polar groups in a POPC bilayer simulated at an area of 64 Å2/lipid matches the distribution of the polar atoms in the closed MscS model. It is consistent with the notion that the route for the transmission of the lateral tension between lipid and protein lies at both the periplasmic and cytoplasmic rims of the channel. The lateral pressure/tension profile is shown in panel D with the red part of the curve representing the tension component. The distributions of lipid groups and pressure across the bilayer are taken from Gullingsrud and Schulten (46).