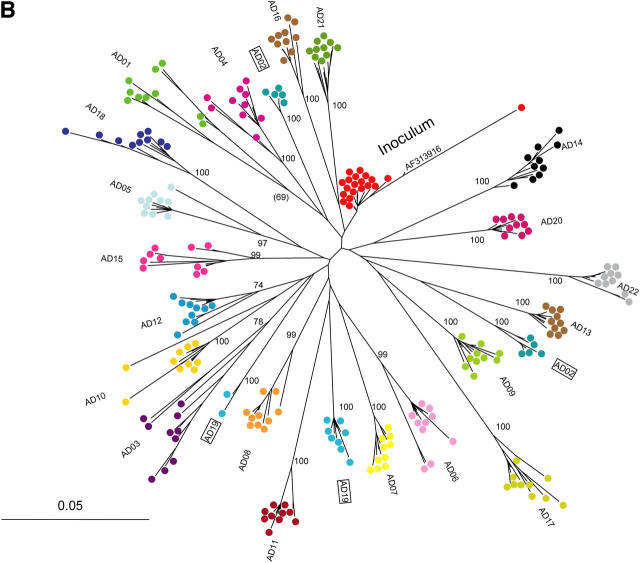
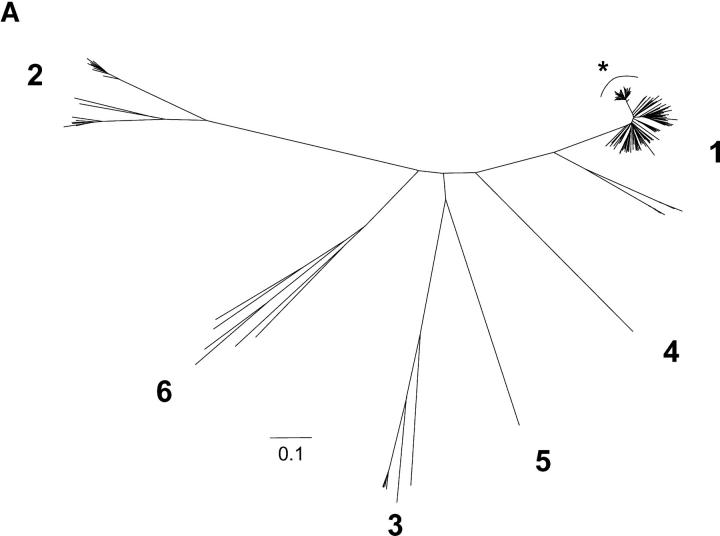
Figure 1.
Phylogenetic analysis of HCV 18–22 yr after common-source outbreak. (A) Phylogenetic tree of 5.2-kb sequence alignment placing outbreak clade (*) in context with reference sequences for all major subtypes. (B) Detailed analysis of the outbreak clade, using 10 cDNA clones from each study subject to obtain the sequence of a 698-nt region spanning the E1–E2 junction. The label “inoculum” indicates 20 clones from inoculum source plasma (10 each from 2 specimens), and a full-length clone (AF313916) obtained in an independent study of this material using smaller amplicons. For both trees, numbers at nodes are bootstrap values, indicating the percentage of 1,000 permuted trees that supported the presence of that node. Bootstrap support was 100% for each of the major clades in A; in B, only bootstrap values >80% are shown, and values for nodes within a study subject's clade were omitted for clarity. Boxes highlight two subjects whose sequences were segregated into two separate clades.