Abstract
Aptamers are single-stranded nucleic acids that fold into defined tertiary structures to bind target molecules with high specificities and affinities. DNA aptamers have garnered much interest as recognition elements for biodetection and diagnostic applications due to their small size, ease of discovery and synthesis, and chemical and thermal stability. Herein, we describe the design and application of a short DNA molecule capable of both protein target binding and amplifiable bioreadout processes. As both recognition and readout capabilities are incorporated into a single DNA molecule, tedious conjugation procedures required for protein-DNA hybrids can be omitted. The DNA aptamer is designed to be amplified directly by either the polymerase chain reaction (PCR) or rolling circle amplification (RCA) processes, taking advantage of real-time amplification monitoring techniques for target detection. A combination of both RCA and PCR provides a wide protein target dynamic range (1 μM to 10 pM).
INTRODUCTION
Aptamers are single-stranded (ss) nucleic acids that are able to fold into defined tertiary structures to bind their targets with high specificities and affinities.(1,2) Targets to which aptamers have successfully been generated against include proteins, small molecules and RNA.(3–8) Aptamers have been suggested as valuable alternatives to antibodies in biodetection and diagnostic applications due to their ease of discovery, their stability, and robust methods for their synthesis.(9–12) A wealth of sensitive aptamer-based biodetection approaches have been reported in which aptamers were labeled with molecules such as redox probes, fluorescent dyes, or nanocrystals to be an integral part of signal transduction.(13–21) In addition, researchers have recently taken advantage of PCR to amplify DNA aptamers for sensitive detection of protein targets. Le and coworkers describe the isolation of a protein-aptamer complex via capillary electrophoresis, followed by PCR amplification of the bound ssDNA and subsequent visualization and quantification by gel electrophoresis.(22) Exonuclease protection of target-bound DNA aptamers has also been used for protein detection,(23) where after hydrolysis of unbound DNA sequences, the bound aptamers were used to template the ligation of a DNA scaffold suitable for real-time (rt) PCR amplification. Another notable technique involved aptamer-coordinated proximity ligation followed by PCR amplification and real-time detection, providing both high sensitivity and selectivity.(24,25) These three approaches exemplify the utility of combining DNA aptamers with PCR for protein target detection but rely on additional steps either before or after PCR for detection.
Detection of proteins using DNA-based amplification techniques was first demonstrated with the advent of immuno-PCR.(26–30) Using antibody-DNA hybrid constructs, the antibody’s binding affinity was complemented by the sensitive detection achievable with PCR. In addition, immuno-DNA detection strategies have been extended to use rolling circle amplification (RCA), an isothermal technique that generates a long ssDNA oligomer tethered to the immuno-DNA conjugate.(31–34) A drawback of these approaches is that the synthesis of the antibody-DNA hybrids can be problematic as controlling the location and number of DNA conjugates per protein is not always straightforward, often leading to heterogeneous ratios of DNA tags per antibody. Recent developments in site-specific conjugation of oligonucleotide tags to proteins using intein chemistry (or chemical ligation) have been very successful,(35) although conjugate preparation remains laborious. A main advantage of solely DNA-based reagents is the possibility of combining both the high affinity of aptamers and amplification techniques for sensitive detection in a single molecular platform, thus reducing synthetic complexity.
Herein, we describe a protein assay approach that uses a short, but yet structurally robust, ssDNA aptamer that is capable of both protein target binding and two complementary amplifiable bioreadout processes (Figure 1A). As both recognition and readout capabilities are incorporated into a single DNA molecule, tedious conjugation procedures required for protein-DNA hybrids are omitted. As for the amplification process, the DNA reagent can be directly subjected to either PCR (27,30) or RCA (31–34) without the need for additional ligation or modification reactions. To demonstrate the approach, we designed a 50nt ssDNA molecule, which was comprised of an aptamer domain, a poly[A]15 linker, and a short 20nt primer (Figure 1B). The 15nt thrombin aptamer was chosen for this study as it has been extensively characterized and binds its target with reasonable affinity (Kd = ~75nM) (36–38) The primer sequence is suitable for RCA amplification, and in conjunction with the recognition domain can prime the PCR reaction. This aptamer-primer hybrid construct effectively binds to the thrombin target protein and can be directly amplified via both PCR and RCA processes.
Figure 1.
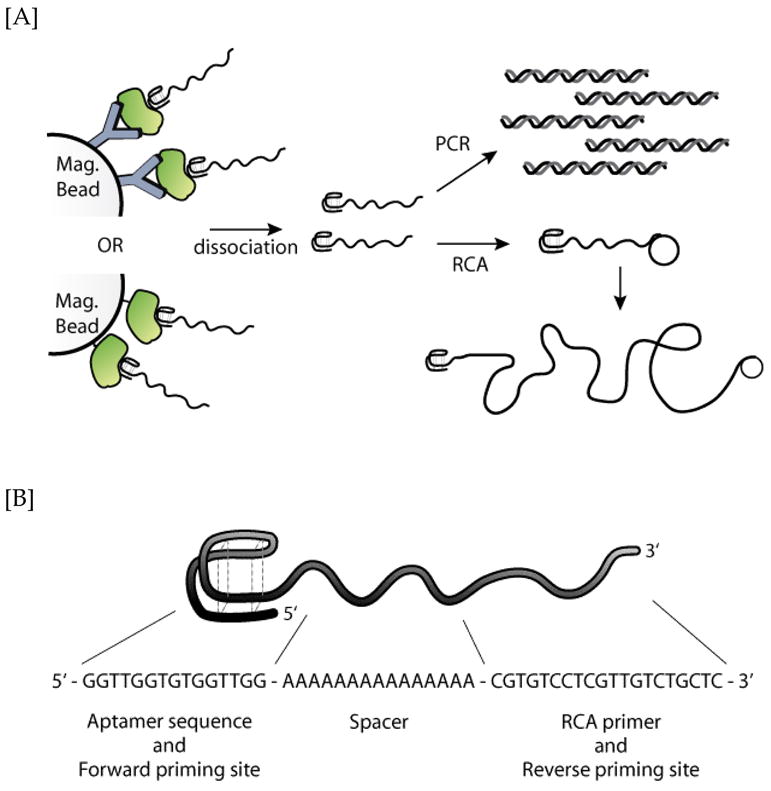
[A] Schematic of the aptamer binding biodetection process. Briefly, either anti-protein antibody or biotinylated protein target was first immobilized onto magnetic beads. After incubation with the DNA aptamers, unbound aptamers were removed. The target-bound aptamer constructs were dissociated and the aptamers were analyzed in real time via either PCR or RCA. [B] Sequence of the 50nt ssDNA aptamer construct, comprised of a thrombin recognition domain, a poly-A linker spacer, and a short RCA primer.
MATERIALS AND METHODS
Materials
Human α-thrombin and human α-thrombin BFPRck (Biotinylated FPR-chloromethylketone) were purchased from Haematologic Technologies Inc. (Essex Junction, VT). Trypsin, Bovine serum albumin (BSA), bovine serum, and polyinosinic–polycytidylic acid potassium salt (dI-dC) were purchased from Sigma (St. Louis, MO). Biotinylated horseradish peroxidase (HRP), EZ biotinylation kit, and EZ biotinylation quantification kit were purchased from Pierce (Rockford, IL). CircLigase, exonuclease I, and RepliPHI phi29 (φ29) DNA polymerase were from Epicentre Biotechnologies (Madison, WI). Deoxynucleotide triphosphates were obtained from New England Biosciences (Ipswich, MA). All oligonucleotides were PAGE purified by the supplier (Integrated DNA Technologies Inc.; Coralville, IA) and used as received. Aptamer: 5′-GGTTGGTGTGGTTGGAAAAAAAAAAAAAAACGTGTCCTCGTTGTCTGCTC –3′ Forward Primer: 5′-GGTTGGTGTGGTTGG-3′ Reverse Primer: 5′-GAGCAGACAACGAGGACACG-3′ RCA template: 5′-GCTTTCGATCGTTCTGAGCAGACAACGAGGACACGCTTACTGAATAGCTA-3′
RCA template circularization
Linear RCA template (1 pmole/μl) was circularized using CircLigase ssDNA ligase at 65°C for 3 hr in reaction buffer containing 50 mM MOPS, pH 7.5, 10 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.05 mM ATP, and ligase (10 units/μl). Uncircularized template was removed by incubating with exonuclease I (0.1 unit/μl) at 37°C for 1 hour, followed by enzyme inactivation at 85°C for 20 min. The circle DNA was used without further purification. Circularization and purity were confirmed by denaturing polyacrylamide gel electrophoresis (15% TBE-Urea Novex gels, Invitrogen; Carlsbad, CA). See Figure S1 for representative gel.
RCA detection
Biotinylated thrombin was immobilized on streptavidin ‘MyOne’ C1 magnetic beads (Invitrogen; Carlsbad, CA) in thrombin binding buffer (1X TBB: 20 mM Tris-acetate pH 7.4, 140 mM NaCl, 5 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 0.02% Tween-20) for 1 hour at room temperature. Biotinylated thrombin immobilization was determined to be ~44 μg per mg of bead by measuring free protein concentration before and after immobilization using the Pierce BCA assay (Rockford, IL). These beads were diluted with uncoated beads to achieve the desired thrombin concentration, so that the final bead concentration was 0.25 mg/ml. The beads were then blocked for two hours with 50 μl blocker solution (1X TBB, 0.1% BSA, 100 μg/ml dI-dC). The aptamer stock solution was heated at 95°C for 3 minutes and added to the blocked beads at a final concentration of 1 μM.
After one hour incubation at room temperature, beads were quickly washed three times with 250 μl of blocker solution (1X TBB, 0.1% BSA and 20 μg/ml dI-dC) and resuspended in 50 μl of deionized water. Samples were then heated at 95°C for 5 minutes to release the bound aptamers. After collection, aptamers (5 μl) were incubated with a 7 μl aliquot of circle DNA (500 nM) in 1X phi29 (φ29) buffer (50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 10 mM (NH4)2SO4, 4mM DTT) for 15 min at 37°C. Remaining RCA components including φ29 DNA polymerase were then added to initiate reaction. Final reaction composition: 15 μl volume, 400 nM circle DNA, 1X φ29 buffer, 200 μg/ml BSA, 1 mM of each dNTP, 40,000 X dilution of SYBR Green I, φ29 (0.5 units/μl). Reactions were monitored in 384-well plates at 31°C with a BMG Labtech FLUOStar (Durham, NC) Optima fluorescence plate reader (λex = 480 nm; λem = 520 nm).
PCR detection
Samples were prepared as above, using an aptamer concentration of 2 nM. After aptamer collection, PCR reactions (20 μl) were prepared with iQ SYBR Green I Supermix (BioRad; Hercules, CA), 0.1 μM of each primer, and 5 μl sample. Real-time PCR was performed using the MyIQ real-time detection system. Cycling parameters were as follows: initial 4 min denaturation at 95°C, 30 cycles of denaturation (95°C, 20 sec), annealing (50.7°C, 20 sec), and elongation (72°C, 30 sec). Fluorescence measurements were acquired after each annealing step. For analysis, a threshold cycle (CT) parameter of 300 relative fluorescence units (RFU) was used. Melt profiles were obtained by monitoring SYBR Green I fluorescence upon heating from 50°C to 95°C at a rate of 2°C/min. Primer dimers were not observed within 30 cycles. Native polyacrylamide gel electrophoresis (4–20% TBE Novex gels, Invitrogen; Carlsbad, CA) were run at 12 mA for 50 min and stained with ethidium bromide. Trypsin (biotinylated following supplier’s procedure) and biotinylated HRP were immobilized on streptavidin ‘MyOne’ C1 magnetic beads and quantified using the Pierce BCA assay. Detection procedures were identical to those used for thrombin.
For sandwich capture assays, magnetic beads functionalized with anti-thrombin antibodies were used. The monoclonal anti-thrombin antibody was biotinylated according to manufacturer’s instructions (Pierce). After purification, the biotinylated antibody was immobilized on streptavidin ‘MyOne’ C1 magnetic beads for 1hr at room temperature, washed, and resuspended in blocking buffer and stored at 4°C until needed. Beads were incubated with various concentration of thrombin diluted in binding buffer. After numerous washing, beads were incubated with aptamers as outlined above.
RESULTS AND DISCUSSION
RCA detection
A few examples demonstrating aptamer-based detection by RCA have been reported recently.(39–42) As mentioned earlier, the 50nt ssDNA aptamer construct employed in this study can be amplified via RCA. Although less sensitive than PCR, the RCA technique is convenient as it can be performed at a constant temperature, e.g. 31°C when φ29 DNA polymerase was used, without the need for thermal cycling. This technique is especially important should a thermocycler not be accessible or practical.
As an initial model system for enzymatic aptamer detection, thrombin protein was directly immobilized on magnetic beads. Thrombin functionalized with a single biotin was immobilized onto ~1 μm diameter streptavidin-coated magnetic beads. The thrombin-labeled beads were then serially diluted into unlabeled beads and subsequently treated with blocker reagents. After incubation with the DNA aptamer, unbound oligonucleotides were removed (details are described in the experimental section). The thrombin-bound aptamers were then dissociated into 50 μl of water by heating the complex at 95°C and 5 μl of the aptamer-containing supernatant were used for RCA analysis. The heat-extracted aptamer was hybridized to a 50nt pre-formed circular ssDNA template on its “built-in” primer tail. Upon the introduction of φ29 DNA polymerase,(43,44) the aptamer’s primer tail was efficiently extended. The DNA extension process can efficiently be monitored in real-time (rt) with SYBR Green I dye.
As seen in Figure 2, the aptamers extracted from the protein-aptamer complex efficiently underwent RCA. Extension of the aptamers at increasing thrombin concentrations showed a corresponding increase in detection signal. At the highest thrombin concentration (1 μM) studied, the bound aptamers were easily detected via the RCA technique in fewer than 10 minutes. The lowest concentration of thrombin protein detected was ~3 nM. From the rt-RCA traces, the individual maximum rates of formation, i.e. the rates of ssDNA extension, were derived for each thrombin concentration. The maximum rates (y-axis) were plotted against the thrombin concentration (x-axis) to yield an estimated limit-of-detection (LoD) of 2 nM. The derived LoD from the RCA amplification technique is ~3 orders of magnitude higher compared to the value derived from PCR (vide infra), which is consistent with the aforementioned limitation in sensitivity for the RCA biodetection approach. Negative controls, consisting of biotinylated trypsin and horseradish peroxidase (HRP), were also performed in the above experimental approach. No RCA amplification was detected in the presence of either negative control targets (Figure 2b), confirming that the observed signal is a result of aptamer binding to the thrombin. Recent advances in rt-RCA detection, such as utilizing sequence-specific hairpin beacons (45) as opposed to the non-specific DNA fluorophore SYBR Green I, could readily be incorporated into our approach to facilitate rt-multiplexed detection. However, for applications where target protein concentrations are not limiting, the RCA approach is an attractive alternative to PCR as the readout process can be performed in real-time under isothermal conditions.
Figure 2.
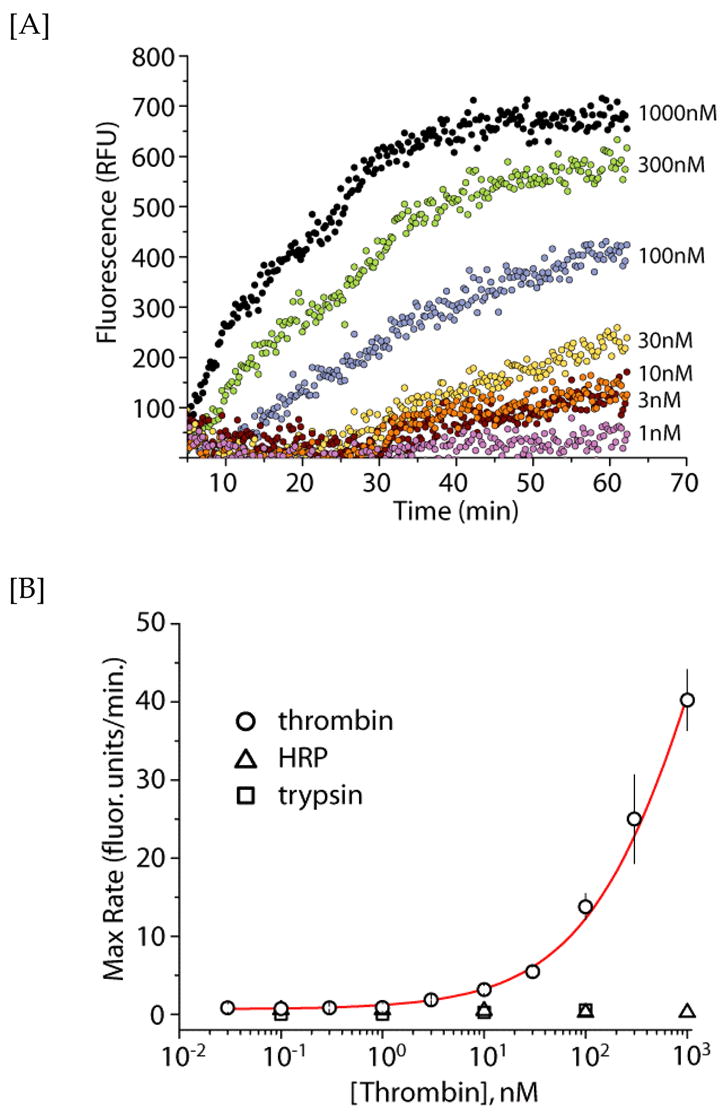
Real-time (rt) RCA detection of thrombin protein target. [A] Representative RCA amplification curves of the aptamer (2 μM) obtained at different thrombin concentrations as a function of time, using SYBR Green I. [B] Detection of thrombin using rt-RCA. The x- and y-axes indicate the thrombin concentration and maximal RCA rate, respectively. The measurements were performed in duplicate and the error bars represent 1 s.d. Negative control experiments indicate no aptamer binding to either HRP (triangles) or trypsin (squares).
PCR detection
The PCR-based amplification assay approach is similar to the aforementioned RCA amplification approach. Briefly, different protein concentrations were first immobilized onto magnetic beads and incubated with the aptamer. After magnetic partitioning of the protein-aptamer complex, the heat-extracted aptamer was amplified by PCR and monitored in real-time with SYBR Green I dye. In Figure 3A, rt-PCR data are presented for the detection of different concentrations of thrombin protein. The increase in fluorescence is relative to the amount of PCR product produced. As expected, at higher concentrations of thrombin, fewer PCR cycles are required for significant signal to accumulate as the concentration of the bound aptamers is higher. The cycle value at which exponential growth of the PCR product occurs (the CT value) is plotted against the thrombin concentration (y- and x-axes, respectively, Figure 3B). An approximate LoD value of ~2 pM can be subsequently derived from this plot. Again for negative controls, no PCR amplified products were detected in the presence of either biotinylated trypsin or HRP (Figure 3B insert), indicating that the observed rt-PCR signal is indeed due to the interaction between the aptamer and its thrombin target.
Figure 3.
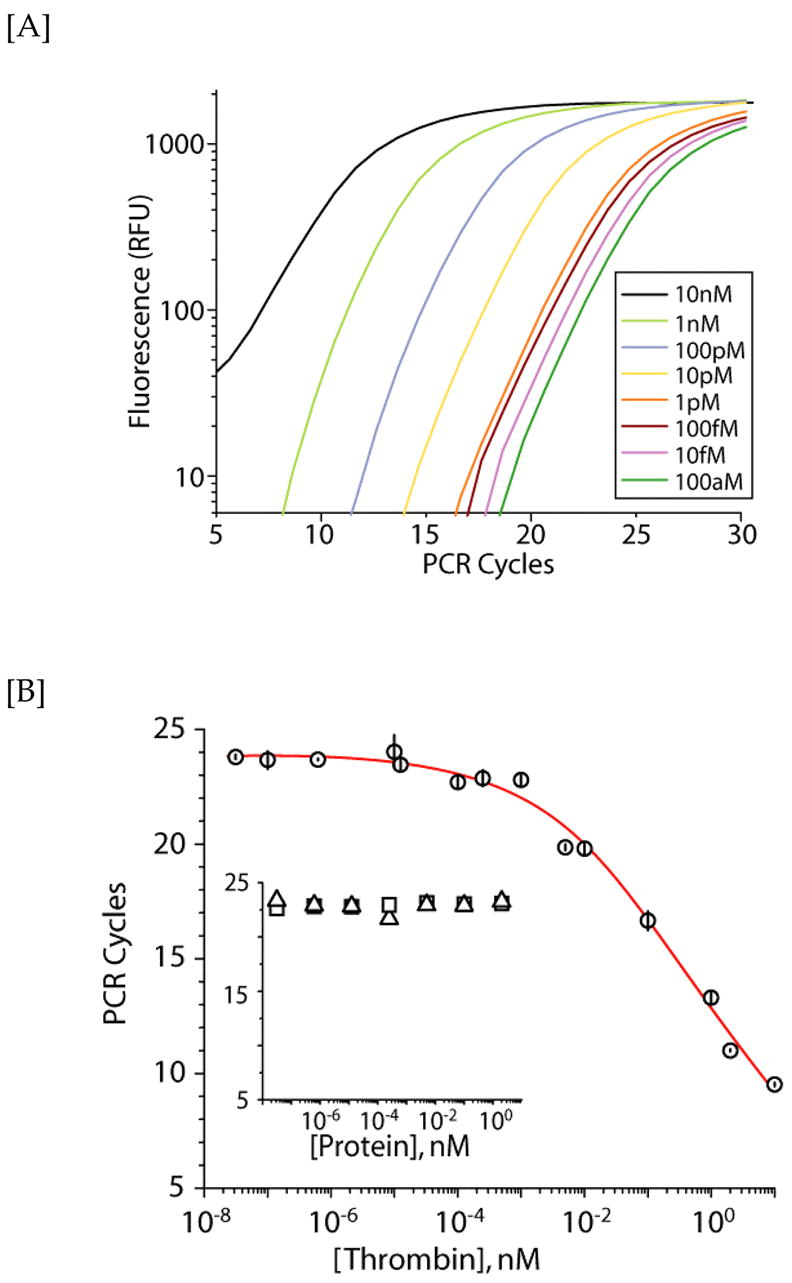
Real-time (rt) PCR detection of thrombin. [A] Representative PCR amplification curves of the aptamer (2 nM) obtained at different thrombin concentrations using SYBR Green I. [B] Detection of thrombin (circles) by rt-PCR using a cycle threshold value of 300 RFU. Data represent two independent experiments performed in triplicate and error bars indicate 1 s.d. Negative control experiments (inset) indicate no aptamer binding to either HRP (triangles) or trypsin (squares).
We observed that there was a linear dependence on protein concentration for the rt- PCR DNA quantification above 1 pM thrombin. However, at lower thrombin concentrations, there is little to no dependence on protein concentration, which is consistent with the hypothesis that non-specific binding of the input DNA aptamer to the magnetic beads may be overwhelming the specific signal from bound aptamers. This observation has been previously reported by Burbulis et al., who used ‘confidence belt’ statistical analyses to extract further information from the non-linear region at the transition between specific and non-specific interactions.(35) Enhanced LoD can also be accomplished by decreasing the amount of aptamer, as a 20-fold reduction in the initial aptamer concentration results in an approximate 20-fold enhancement in the LoD (to ~100 fM; see supplementary data section Figure S2). Conversely, increasing the concentration of aptamer above 2 nM resulted in a concomitant reduction in sensitivity (data not shown). Our studies also underscore the importance of blocking and washing parameters, as well as the choice of magnetic bead platform. For instance, initial optimization of these parameters demonstrated that blocking buffer supplemented with the polyanionic dI-dC and 0.1% BSA provided 2- to 5-fold greater sensitivity. Furthermore, wash buffers incorporating these blocking agents were more effective than buffer alone, whereas increasing the number of washes beyond three was less important in overall sensitivity. Using small polymer-coated magnetic beads (1 μm, Dynabeads MyOne Streptavidin, Invitrogen) also provided a ca. 4-fold enhancement in the LoD when compared to larger polystyrene magnetic beads (2.8 μm, Dynabeads M-270 Streptavidin, Invitrogen). These observations demonstrate a certain degree of control over the LoD, which, alongside the design of detection platform and assay parameters, can be tailored for each specific desired application.
To confirm that the rt-PCR amplified product was indeed the input aptamer, the double-stranded (ds) DNA products were analyzed using gel electrophoresis. A single band of 50bp dsDNA was consistently obtained at all protein concentrations investigated (see supplementary data, Figure S3A). To further corroborate this observation, the amplified dsDNA was also subjected to a DNA-melt analysis, which demonstrated that all the rt-PCR amplified dsDNA have a single melting profile (Tm = ~75°C), consistent with the presence of only a single dsDNA PCR product (Figure S3B).
Detection of biotinylated thrombin protein target on magnetic beads provided a model system for our initial experiments. To demonstrate real-world applicability of our systems, thrombin was also captured from solution in a sandwich assay format via magnetic beads functionalized with an anti-thrombin antibody (Ab). The beads were subsequently incubated with the 50nt input aptamer and detected by rt-PCR. As seen in Figure 4A, low picomolar concentrations could be detected by rt-PCR. The data obtained from the sandwich capture assay are in accord with the detection of thrombin directly conjugated to beads.
Figure 4.
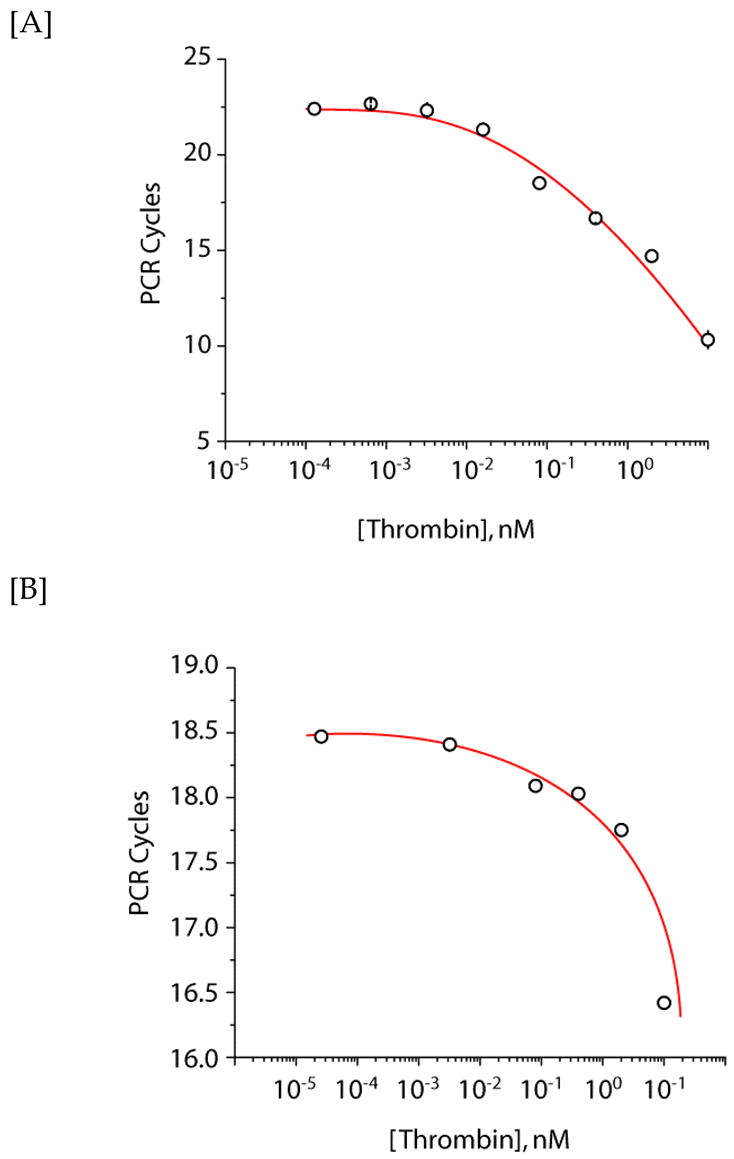
Thrombin capture and real-time (rt) PCR detection. [A] Serial dilutions of thrombin were incubated with magnetic beads functionalized with an anti-thrombin antibody. Aptamers (2 nM) were subsequently added and detected by rt-PCR using a cycle threshold value of 300 RFU. [B] rt-PCR detection of thrombin in 10% bovine serum (v/v in binding buffer). Thrombin was diluted into 10% bovine serum, and incubated with Ab-functionalized beads. After briefly washing the beads, the aptamer (2 nM) was introduced and subsequently incubated for 1 hr. The beads were isolated and heat-dissociated aptamers were then detected by rt-PCR using a cycle threshold value of 300 RFU.
Last, accurate and quantitative detection of protein targets in complex mixture is highly desired in all biological immunoassays. To investigate if the rt-PCR technique could be used to detect the target thrombin protein in complex mixtures, the sandwich capture assay was also performed in bovine serum (Figure 4B). We have chosen to dilute the thrombin target in 10% serum (v/v in binding buffer), in which the anticipated dose response was again observed. This indicates that specific capture and detection of thrombin in serum is possible. However, a reduction in sensitivity was observed when performed in 10% serum, with the single-to-noise significantly diminishing to undetectable levels above 10% serum. This serum dependence is presumably due to: (i) non-specific binding of the serum proteins to the anti-thrombin Ab coated magnetic beads, resulting in subsequent decreased binding of thrombin to the Ab, and (ii) increased non-specific binding of the aptamers to the various serum components in solution, effectively depleting the pool of aptamers available for target binding. It is also likely that the non-specific binding of serum proteins to the magnetic beads results in the increased nonspecific binding of the DNA hybrid, a complication which is not unique to our approach. We anticipate that the confounding effects of serum will be similar to other clinically relevant matrices. General solutions to this are optimization of blocking and washing conditions. Alternatively, aptamers can be selected under conditions that mimic those anticipated during detection. Consequently, these aptamers may present greater specificity toward the target in complex media. Nonetheless, these experiments indicate that the DNA hybrid and rt-PCR assay are sufficiently robust to tolerate detection of ca. 10 pM thrombin in 10% bovine serum.
CONCLUSION
We have demonstrated a simple, yet efficient, amplification assay procedure based on a single oligonucleotide DNA aptamer platform for sensitive detection of protein target leveraging real-time amplification monitoring instrumentation. The aptamer employed was able to simultaneously function as both affinity ligand and reporter group for the thrombin target protein. The 50nt ssDNA aptamer sequence itself can be used directly in PCR amplification without additional ligation or modification steps, exhibiting a low picomolar LoD for thrombin, even in 10% serum. Additionally, since the aptamer already incorporates an appropriate RCA primer, it can also be detected at isothermal conditions by RCA. This approach is especially useful should a thermocycler not be readily available. The entire assay spans a wide protein target dynamic range, accommodating concentrations from 1 μM down to low pM. As no detrimental effect on native aptamer affinity was observed in the presence of the linker and primer domains (data not shown), we anticipate that this approach would be applicable to a wide variety of aptamers given proper attention to both the design and testing of the aptamer. As a result, a diverse range of aptamer-binding targets, e.g. proteins and cells, could be accommodated with this biodetection assay. With the employment of magnetic beads as the partitioning platform, the described approach should readily be amendable for multiplexed detection using a microfluidic, high-throughput assay platform and be fully automated. In addition, RCA amplification of aptamer-hybrids can be readily adapted for protein detection in chip-based applications as the signal is spatially localized at the site of the binding event. (33)
Supplementary Material
Acknowledgments
This work was performed under the auspices of the U.S. Department of Energy by the University of California, Lawrence Livermore National Laboratory, under contract W-7405-Eng-48. N.O.F. acknowledges CMLS for a directorate postdoctoral fellowship and J.B.T. acknowledges the partial support of NIH Grant AI065359.
Abbreviations
- RCA
rolling circle amplification
- PCR
polymerase chain reaction
- nt
nucleotides
- rt
real-time
- Tm
melting temperature
- LoD
limit of detection
- RFU
Relative Fluorescence Units
- ss
single-stranded
- ds
double-stranded
- Ab
antibody
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 2.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 3.Ellington AD, Szostak JW. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature. 1992;355:850–852. doi: 10.1038/355850a0. [DOI] [PubMed] [Google Scholar]
- 4.Huizenga DE, Szostak JW. A DNA aptamer that binds adenosine and ATP. Biochemistry. 1995;34:656–665. doi: 10.1021/bi00002a033. [DOI] [PubMed] [Google Scholar]
- 5.Koizumi M, Breaker RR. Molecular recognition of cAMP by an RNA aptamer. Biochemistry. 2000;39:8983–8992. doi: 10.1021/bi000149n. [DOI] [PubMed] [Google Scholar]
- 6.Lauhon C, Szostak J. Invitro Selection of Rna Molecules That Specifically Bind Riboflavin. Faseb Journal. 1993;7:A1087–A1087. [Google Scholar]
- 7.Tok JB, Cho J, Rando RR. RNA aptamers that specifically bind to a 16S ribosomal RNA decoding region construct. Nucleic Acids Res. 2000;28:2902–2910. doi: 10.1093/nar/28.15.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tarasow TM, Tarasow SL, Eaton BE. RNA-catalysed carbon-carbon bond formation. Nature. 1997;389:54–57. doi: 10.1038/37950. [DOI] [PubMed] [Google Scholar]
- 9.Jayasena SD. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 10.Bunka DH, Stockley PG. Aptamers come of age - at last. Nat Rev Microbiol. 2006;4:588–596. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- 11.Wilson DS, Szostak JW. In vitro selection of functional nucleic acids. Annu Rev Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 12.Porschewski P, Grattinger MA, Klenzke K, Erpenbach A, Blind MR, Schafer F. Using aptamers as capture reagents in bead-based assay systems for diagnostics and hit identification. J Biomol Screen. 2006;11:773–781. doi: 10.1177/1087057106292138. [DOI] [PubMed] [Google Scholar]
- 13.Baker BR, Lai RY, Wood MS, Doctor EH, Heeger AJ, Plaxco KW. An electronic, aptamer-based small-molecule sensor for the rapid, label-free detection of cocaine in adulterated samples and biological fluids. J Am Chem Soc. 2006;128:3138–3139. doi: 10.1021/ja056957p. [DOI] [PubMed] [Google Scholar]
- 14.Nutiu R, Li Y. Structure-switching signaling aptamers. J Am Chem Soc. 2003;125:4771–4778. doi: 10.1021/ja028962o. [DOI] [PubMed] [Google Scholar]
- 15.Nutiu R, Li Y. Aptamers with fluorescence-signaling properties. Methods. 2005;37:16–25. doi: 10.1016/j.ymeth.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Pavlov V, Shlyahovsky B, Willner I. Fluorescence detection of DNA by the catalytic activation of an aptamer/thrombin complex. J Am Chem Soc. 2005;127:6522–6523. doi: 10.1021/ja050678k. [DOI] [PubMed] [Google Scholar]
- 17.Pavlov V, Xiao Y, Shlyahovsky B, Willner I. Aptamer-functionalized Au nanoparticles for the amplified optical detection of thrombin. J Am Chem Soc. 2004;126:11768–11769. doi: 10.1021/ja046970u. [DOI] [PubMed] [Google Scholar]
- 18.Levy M, Cater SF, Ellington AD. Quantum-dot aptamer beacons for the detection of proteins. Chembiochem. 2005;6:2163–2166. doi: 10.1002/cbic.200500218. [DOI] [PubMed] [Google Scholar]
- 19.Chu TC, Shieh F, Lavery LA, Levy M, Richards-Kortum R, Korgel BA, Ellington AD. Labeling tumor cells with fluorescent nanocrystal-aptamer bioconjugates. Biosens Bioelectron. 2006;21:1859–1866. doi: 10.1016/j.bios.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 20.Xiao Y, Piorek BD, Plaxco KW, Heeger AJ. A reagentless signal-on architecture for electronic, aptamer-based sensors via target-induced strand displacement. J Am Chem Soc. 2005;127:17990–17991. doi: 10.1021/ja056555h. [DOI] [PubMed] [Google Scholar]
- 21.Xiao Y, Lubin AA, Heeger AJ, Plaxco KW. Label-free electronic detection of thrombin in blood serum by using an aptamer-based sensor. Angew Chem Int Ed Engl. 2005;44:5456–5459. doi: 10.1002/anie.200500989. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Wang Z, Li XF, Le XC. Ultrasensitive detection of proteins by amplification of affinity aptamers. Angew Chem Int Ed Engl. 2006;45:1576–1580. doi: 10.1002/anie.200503345. [DOI] [PubMed] [Google Scholar]
- 23.Wang XL, Li F, Su YH, Sun X, Li XB, Schluesener HJ, Tang F, Xu SQ. Ultrasensitive detection of protein using an aptamer-based exonuclease protection assay. Anal Chem. 2004;76:5605–5610. doi: 10.1021/ac0494228. [DOI] [PubMed] [Google Scholar]
- 24.Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gustafsdottir SM, Ostman A, Landegren U. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol. 2002;20:473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- 25.Gustafsdottir SM, Schallmeiner E, Fredriksson S, Gullberg M, Soderberg O, Jarvius M, Jarvius J, Howell M, Landegren U. Proximity ligation assays for sensitive and specific protein analyses. Anal Biochem. 2005;345:2–9. doi: 10.1016/j.ab.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 26.Sano T, Smith CL, Cantor CR. Immuno-PCR: very sensitive antigen detection by means of specific antibody-DNA conjugates. Science. 1992;258:120–122. doi: 10.1126/science.1439758. [DOI] [PubMed] [Google Scholar]
- 27.Niemeyer CM, Adler M, Wacker R. Immuno-PCR: high sensitivity detection of proteins by nucleic acid amplification. Trends Biotechnol. 2005;23:208–216. doi: 10.1016/j.tibtech.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 28.Adler M, Wacker R, Niemeyer CM. A real-time immuno-PCR assay for routine ultrasensitive quantification of proteins. Biochem Biophys Res Commun. 2003;308:240–250. doi: 10.1016/s0006-291x(03)01364-0. [DOI] [PubMed] [Google Scholar]
- 29.Allen RC, Rogelj S, Cordova SE, Kieft TL. An immuno-PCR method for detecting Bacillus thuringiensis Cry1Ac toxin. J Immunol Methods. 2006;308:109–115. doi: 10.1016/j.jim.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 30.Niemeyer CM, Wacker R, Adler M. Combination of DNA-directed immobilization and immuno-PCR: very sensitive antigen detection by means of self-assembled DNA-protein conjugates. Nucleic Acids Res. 2003;31:e90. doi: 10.1093/nar/gng090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gusev Y, Sparkowski J, Raghunathan A, Ferguson H, Jr, Montano J, Bogdan N, Schweitzer B, Wiltshire S, Kingsmore SF, Maltzman W, Wheeler V. Rolling circle amplification: a new approach to increase sensitivity for immunohistochemistry and flow cytometry. Am J Pathol. 2001;159:63–69. doi: 10.1016/S0002-9440(10)61674-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nilsson M, Gullberg M, Dahl F, Szuhai K, Raap AK. Real-time monitoring of rolling-circle amplification using a modified molecular beacon design. Nucleic Acids Res. 2002;30:e66. doi: 10.1093/nar/gnf065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schweitzer B, Roberts S, Grimwade B, Shao W, Wang M, Fu Q, Shu Q, Laroche I, Zhou Z, Tchernev VT, Christiansen J, Velleca M, Kingsmore SF. Multiplexed protein profiling on microarrays by rolling-circle amplification. Nat Biotechnol. 2002;20:359–365. doi: 10.1038/nbt0402-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong XB, Lizardi PM, Huang XH, Bray-Ward PL, Ward DC. Visualization of oligonucleotide probes and point mutations in interphase nuclei and DNA fibers using rolling circle DNA amplification. Proc Natl Acad Sci U S A. 2001;98:3940–3945. doi: 10.1073/pnas.061026198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burbulis I, Yamaguchi K, Gordon A, Carlson R, Brent R. Using protein-DNA chimeras to detect and count small numbers of molecules. Nat Methods. 2005;2:31–37. doi: 10.1038/nmeth729. [DOI] [PubMed] [Google Scholar]
- 36.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 37.Macaya RF, Schultze P, Smith FW, Roe JA, Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc Natl Acad Sci U S A. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tasset DM, Kubik MF, Steiner W. Oligonucleotide inhibitors of human thrombin that bind distinct epitopes. J Mol Biol. 1997;272:688–698. doi: 10.1006/jmbi.1997.1275. [DOI] [PubMed] [Google Scholar]
- 39.Cho EJ, Yang L, Levy M, Ellington AD. Using a deoxyribozyme ligase and rolling circle amplification to detect a non-nucleic acid analyte, ATP. J Am Chem Soc. 2005;127:2022–2023. doi: 10.1021/ja043490u. [DOI] [PubMed] [Google Scholar]
- 40.Di Giusto DA, Wlassoff WA, Gooding JJ, Messerle BA, King GC. Proximity extension of circular DNA aptamers with real-time protein detection. Nucleic Acids Res. 2005;33:e64. doi: 10.1093/nar/gni063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jarvius J, Melin J, Goransson J, Stenberg J, Fredriksson S, Gonzalez-Rey C, Bertilsson S, Nilsson M. Digital quantification using amplified single-molecule detection. Nat Methods. 2006;3:725–727. doi: 10.1038/nmeth916. [DOI] [PubMed] [Google Scholar]
- 42.Yang L, Fung CW, Cho EJ, Ellington AD. Real-time rolling circle amplification for protein detection. Anal Chem. 2007;79:3320–3329. doi: 10.1021/ac062186b. [DOI] [PubMed] [Google Scholar]
- 43.Soengas MS, Gutierrez C, Salas M. Helix-destabilizing activity of phi 29 single-stranded DNA binding protein: effect on the elongation rate during strand displacement DNA replication. J Mol Biol. 1995;253:517–529. doi: 10.1006/jmbi.1995.0570. [DOI] [PubMed] [Google Scholar]
- 44.Blanco L, Bernad A, Lazaro JM, Martin G, Garmendia C, Salas M. Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. J Biol Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]
- 45.Smolina IV, Demidov VV, Cantor CR, Broude NE. Real-time monitoring of branched rolling-circle DNA amplification with peptide nucleic acid beacon. Anal Biochem. 2004;335:326–329. doi: 10.1016/j.ab.2004.07.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


