Abstract
In severe combined immunodeficient (scid) mice, V(D)J recombination is severely impaired due to a recessive mutation (scid). Thus, we were surprised to find in this study that Vλ1–Jλ1 rearrangement is routinely detectable in scid fetal liver, adult bone marrow, and spleen in the apparent absence of completed VH–DJH and Vκ–Jκ rearrangements. Particularly surprising, we found the level of Vλ1–Jλ1 rearrangement in scid fetal liver to be comparable to that in fetal liver of wild-type mice. The majority of scid Vλ1–Jλ1 rearrangements contained abnormal deletions at the VJ junction, consistent with the known effect of scid. However, ∼15% of Vλ1–Jλ1 rearrangements lacked abnormal deletions. Productive λ1 transcripts resulting from in-frame rearrangements were readily detectable in scid adult bone marrow and spleen, consistent with our ability to detect λ1-expressing cells by flow cytometry in the spleens of bcl-2–transgenic scid mice. Strikingly, λ1 transcripts from individual scid mice often showed VJ junctional sequences with the same recurring palindromic (P) additions of three, four, or five nucleotides. To account for these findings, we suggest that (a) nonhomologous end joining of Vλ1 and Jλ1 coding ends in fetal B lineage cells may not be (severely) impaired by scid; (b) recurring P additions in scid λ1 transcripts may reflect certain molecular constraints imposed by scid on the resolution of Vλ1 and Jλ1 hairpin coding ends; and (c), scid lymphocytes with productively rearranged Vλ1 and Jλ1 elements may differentiate into recombinase-inactive cells and emigrate from bone marrow to spleen.
Keywords: B cell differentiation, pro-B cells, premature Igλ recombination, VJλ junctional diversity, P additions
Introduction
The rearrangement of Ig genes proceeds in an ordered fashion (for review see references 1 and 2). It begins with rearrangement of the H chain gene elements, DH and JH, followed by VH–DJH rearrangement 3. H chain gene rearrangement is initiated at the pro-B cell stage and is generally followed by L chain gene rearrangement at the pre-B cell stage 4 5 6, with Vλ–Jλ rearrangement occurring later 7 8 9 10 and/or less frequently 11 12 13 than Vκ–Jκ rearrangement. The order of H and L chain gene rearrangement is not absolute, however, as studies with B lineage cell lines 14 15 16 and sorted pro-B cells 5 6 indicate that rearrangement at the κ or λ locus may precede or occur independently of H chain gene rearrangement. Also, inactivation of the κ locus by gene targeting has shown that rearrangement of the λ locus does not require prior rearrangement of the κ locus 17 18, consistent with earlier evidence for independent rearrangement of λ and κ loci in various cell lines 16 19 20 21 and in κ-transgenic mice 22. Independent rearrangement of λ and κ loci is in agreement with a stochastic model of L chain gene rearrangement 11 23 and contrary to a strictly regulated model of L chain gene rearrangement (for review see reference 2).
Consistent with the stochastic model of L chain gene rearrangement, we report here a low frequency of Vλ1–Jλ1 rearrangement in severe combined immunodeficient (scid) and in wild-type (wt) mice with a targeted deletion of the JH locus (JHT mice) 24. As JHT mice lack a functional H chain locus, Vλ1–Jλ1 rearrangement in these mice must occur independently of H chain rearrangement. Furthermore, as both scid and JHT mice lack pre-B cells, the observed Vλ1–Jλ1 rearrangement is inferred to occur at the pro-B cell stage and independently of Vκ–Jκ rearrangement.
Although a low frequency of Vλ1–Jλ1 rearrangement at the pro-B cell stage in JHT mice could have been predicted based on the stochastic model of L chain gene rearrangement, the regular occurrence of such rearrangement in scid pro-B cells would not have been predicted. In scid mice, V(D)J recombination is severely impaired as a result of a DNA repair defect 25 26 27. The defect is due to a nonsense mutation in the gene coding for the catalytic subunit of DNA protein kinase (DNA-PKcs) 28 29 30. Because of this mutation, developing scid lymphocytes cannot efficiently join V, D, and J coding ends resulting from the initiation of V(D)J recombination 31 32 33 34. Consequently, most developing scid lymphocytes are thought to die prematurely with persisting DNA breaks. Therefore, given that Vλ1–Jλ1 rearrangement generally follows DH–JH and VH–DJH rearrangement and is much less frequent than Vκ–Jκ rearrangement, we would not expect to detect Vλ1–Jλ1 rearrangement in scid mice.
Nonetheless, as shown here, Vλ1–Jλ1 rearrangement is routinely detectable in fetal liver, adult bone marrow, and spleen of individual scid mice. Moreover, in scid fetal liver we found the level of Vλ1–Jλ1 rearrangement to be comparable to that in wt fetal liver. Although most scid Vλ1–Jλ1 rearrangements showed abnormal deletions of Vλ1 and/or Jλ1 nucleotides at the VJ junction, ∼15% lacked abnormal deletions. Productive λ1 transcripts resulting from in-frame Vλ1–Jλ1 rearrangements were clearly evident in bone marrow and spleens of individual scid mice. Most scid λ1 transcripts displayed the same recurring palindromic (P) additions of three, four, or five nucleotides. P addition 35 36 is thought to result from asymmetric nicking of a hairpin coding end 37 38 39 40, followed by fill-in of the overhang and joining to another coding end.
Our findings raise several puzzling issues: (a) Why is the level of Vλ1–Jλ1 rearrangement comparable in fetal liver of scid and wt mice; (b) Why do most λ1 transcripts of individual scid mice show the same recurring P additions; and (c) How do λ1 expressing scid cells survive the deleterious effect of scid and apparently emigrate from bone marrow to spleen? These three issues are discussed.
Materials and Methods
Mice.
C.B-17 mice homozygous and heterozygous for the scid mutation 41 are here denoted as scid and scid/+ mice, respectively. C.B-17 scid mice hemizygous for the bcl-2-36 transgene (bcl-2 scid mice; reference 42) were obtained from S. Cory (The Walter and Eliza Hall of Medical Research, Melbourne, Australia). Genotyping of bcl-2 mice was done by PCR using DNA from tail snips 43 and oligonucleotide primers for the SV40 sequence included in the transgene 44. Mice with both of their recombination activation gene (RAG)1 loci inactivated by gene targeting (RAG−/− mice; reference 45) and mice with their JH elements deleted by gene targeting 24 were provided by R. Hardy (Fox Chase Cancer Center). The targeted JHT allele was backcrossed onto C.B-17 mice for three backcross generations (N3). N3F1 mice were intercrossed to generate N3F2 mice homozygous for the JHT allele (JHT mice). JHT mice were crossed with scid mice to obtain JHT/+, scid/+ mice; these were then intercrossed to obtain JHT scid mice. Genotyping for the wt and inactivated JH allele was done by PCR using tail DNA and primers specific for the wt and inactivated JH locus (see JH1 and JHT oligonucleotides below). All of the above mice were bred and maintained at the Fox Chase Cancer Center and were analyzed between the ages of 6 and 12 wk.
Flow Cytometric Analysis.
Flow cytometry was used to test for the presence of cells with surface Igλ1 (λ1+ cells) in scid and bcl-2 scid mice. In brief, spleen cells of individual scid, scid/+, bcl-2 scid, bcl-2 scid/+, and RAG1−/− mice were stained with biotin-conjugated anti-CD8 (53.6), allophycocyanin (PharMingen)-conjugated anti-CD45 (B220), and FITC-conjugated anti-λ1 (R11-153–FITC; PharMingen) in the manner previously described 46. Cells were analyzed by three-color flow cytometry using a dual laser FACStarPLUS™ (Becton Dickinson). Binding of biotinylated antibodies was revealed by Texas Red–conjugated streptavidin (Southern Biotechnology). Dead cells were identified by propidium iodide staining and excluded from analysis. Gates were set to score λ1+ cells based on the distribution of λ1 staining of spleen cells in the scid/+ positive controls. Due to the paucity of λ1+ cells in scid and bcl-2 scid mice, between 0.5 and 1.0 × 107 spleen cells were analyzed per mouse. Cells were simultaneously stained for the B and T specific markers B220 and CD8, respectively, to ensure that cells scored as λ1+ were indeed B lineage cells (i.e., λ1+B220+CD8−). Spleen cells from RAG1−/− mice served as negative controls for background staining of λ1.
Oligonucleotides.
Oligonucleotides were synthesized by an Applied Biosystems 394 DNA/RNA Synthesizer. Oligonucleotides used as primers for PCR or reverse transcriptase (RT)-PCR were as follows: VH (#91), 5′-GCCGGATCCGTGCAGCTGGTGGAGTCTGG-3′; DH (#285), 5′-ACTGCTACCTCTGGCCCCACCAG-3′; JH4 (#361R), 5′-AGATAATCTGTCCTAAAGGCTC-3′; Cμ (#289), 5′-ATGCAGATCTCTGTTTTTGCC-TCC-3′; Vκ (#68), 5′-GGCTGCAGGACATTGTGCTGACCCAATCTCCAGCTTCT-3′; Jκ2 (#367), 5′-GGTAGACAATTATCCCTCTTCCCCTAGT-3′; Cκ (#130), 5′-ATGGATCCAGTTGGTGCAGCATC-3′; Vλ1ext (#282), 5′-TCTCCT-GGCTCTCAGCTCAG-3′; Vλ1int (#294), 5′-AGGAATCTGCACTCACCACATC-3′; Jλ1 (#271), 5′-GCACCTCAAGTC-TTGGAGAG-3′; Cλ1 (#283), 5′-GAGGAAGGTGGAAACAG-GGTG-3′; β2ML (#229), 5′-GAATGGGAAGCCGAACATAC-TGAACTG-3′; β2MR (#230), 5′-TGCTGATCACATGTCTC-GATCC-3′; SV40L (#355), 5′-GGAACTGATGAATGGGAGC-AGTGG-3′; SV40R (#356), 5′-GCAGACACTCTATGCCTGTG-3′; JHTL (#370), 5′-CCTTGCGCAGCTGTGCTCGACGTTG-3′; JHTR (#371), 5′-GCCGCATTGCATCAGCCATGATGGA-3′; JH1L (#368), 5′-GGACCAGGGGGCTCAGGTC-ACTCAGG-3′; and JH1R (#369), 5′-GAGGAGACGGTGACCGTGGTGCCTGC-3′.
Genomic PCR.
∼5 × 106 bone marrow or spleen cells were used to prepare genomic DNA by PureGene Kit (Gentra Systems). PCR was carried out in 50 μl with ∼106 cell genome equivalents of DNA. Controls for nonspecific amplification of PCR products included the use of RAG−/− DNA and no DNA template in the reaction. Reactants included oligonucleotide primers for DH and JH, VH and JH, Vκ and Jκ, Vλ1 and Jλ1 elements or for the β2 microglobulin (β2M) locus along with 220 μM each of dATP, dGTP, dCTP, and dTTP, 0.4 μM primers, 20 μM Tris-HCl, pH 8.4, 50 μM KCl, 1.5 μM MgCl2, and 2.5 U of AmpliTaq DNA polymerase. The cycling reaction consisted of an initial denaturation for 4 min at 95°C, with 23 cycles of 1 min at 94°C, 45 s at 68°C, and 1 min at 72°C, and a final elongation step for 5 min at 72°C. 1/10 of each PCR reaction (1/20 for β2M control) was electrophoresed through 1.5% LE agarose (FMC Bioproducts) in 1× Tris-acetate-EDTA buffer, turboblotted by alkaline transfer onto maximum strength Nytran Plus membranes (Schleicher & Schuell) and hybridized in Denhardt's solution with the appropriate probes. Radioactive α-[32P]dCTP labeling was done by random priming using the Prime-It II Kit (Stratagene). Hybridization probes included pJH6.3 47, pECκ 11, and a PCR-amplified and gel-purified (QiaexII; Qiagen) VJλ1 gene fragment to score for DH–JH (or VH–DJH), Vκ–Jκ, and Vλ1–Jλ1 rearrangements, respectively. As a control for the amount of input DNA, a portion of the nonrearranging β2M gene was PCR amplified and hybridized to a β2M-specific probe. Blots were exposed to X-Omat (Eastman Kodak Co.) autoradiographic film and also to a PhosphorImaging plate for quantitation by a BAS1000Mac Bio-Imaging Analyzer (Fuji Photo Film Co.).
RT-PCR.
Total RNA from ∼5 × 106 bone marrow or spleen cells was obtained by using RNEasy (Qiagen) as prescribed by the manufacturer. RNA was eluted into DEPC-treated H2O and stored at −73°C. RNA from the equivalent of ∼1.5 × 106 bone marrow cells or ∼3.0 × 106 spleen cells was used to synthesize first strand cDNA using SuperscriptII RT and 100 ng of random hexamers (Amersham Pharmacia Biotech) as directed by the manufacturer (GIBCO BRL). A portion of this cDNA (equivalent to ∼3 × 105 bone marrow cells or ∼6 × 105 spleen cells) was amplified by PCR using 220 μM each of dATP, dGTP, dCTP, and dTTP, 0.4 μM primers, 20 μM Tris-HCl, pH 8.4, 50 μM KCl, 1.5 μM MgCl2, and 2.5 U of AmpliTaq DNA polymerase (PerkinElmer) in a reaction volume of 50 μl. Controls for nonspecific amplification of PCR products included RAG−/− cDNA and no cDNA template in the reaction. Semiquantitative PCR using a PTC-100 Thermal Controller (MJ Research) was carried out after an initial denaturation for 4 min at 95°C, with 23 cycles of 1 min at 94°C, 45 s at 65°C, and 1 min at 72°C, and a final elongation step for 5 min at 72°C. Southern blotting and hybridization was carried out as described above. The hybridization probes included pCμ3741 48, pECκ, and gel-purified VJλ1 and β2M PCR-amplified gene fragments.
Quantitation.
Conditions for semiquantitative PCR were determined by varying cycle number and the amount of input DNA (or cDNA). Filters were exposed to a Fuji imaging plate to quantify the amount of α-32P–hybridized probe in experimental samples relative to that in control (reference) samples using a BAS1000Mac Bio-Imaging Analyzer (Fuji Photo Film Co.). We found that with 23 cycles of amplification, the amount of PCR product was proportional to the amount of input DNA (from 106 cells) at several different dilutions. Similarly, at 23 cycles, the amount of RT-PCR product was found to be proportional to the amount of input cDNA (from 3–6 × 105 cells) at several different dilutions.
Sequence Analysis.
To ensure sufficient PCR product for cloning, one microliter from the primary PCR or RT-PCR reaction was subjected to an additional 15 cycles of PCR using conditions as above. The Vλlint primer was used with Jλ1 or Cλ1 for recovery of junctional sequences from genomic DNA or cDNA, respectively. PCR products were electrophoresed through 1.5% LE agarose, purified using QiaexII, and cloned into pCR2.1 for transformation of INVαF′ bacteria (Invitrogen). Recombinant colonies were randomly chosen for plasmid recovery by Perfect Prep (5Prime-3Prime, Inc.). Plasmids were submitted for cycle sequencing using the ABI Prism Dye Terminator Reaction Kit and an ABI 377 DNA Sequencer (PerkinElmer).
Results
Evidence for Vλ1–Jλ1 Rearrangement before the Pre-B cell Stage.
To test whether the λ1 locus can rearrange early in B cell differentiation, we assayed for the presence of nongermline λ1 transcripts in scid mice and also in bcl-2 scid mice. As shown in Fig. 1 A, λ1 transcripts resulting from Vλ1–Jλ1 rearrangement (λ1 transcripts) were clearly evident in the bone marrow of scid mice and more so in the bone marrow of bcl-2 scid mice. The higher abundance of λ1 transcripts in the latter mice presumably reflects the greater longevity of B lineage cells in bcl-2 scid mice than in scid mice 42. Fig. 1 A also illustrates that transcripts resulting from Vκ–Jκ rearrangement (κ transcripts) were routinely detectable in bone marrow of bcl-2 scid but not scid mice, whereas transcripts resulting from VH–DJH rearrangement (μ transcripts) were barely detectable in some bcl-2 scid mice and not at all in scid mice (Fig. 1 A). Consistent with previous reports of detectable DH–JH rearrangement in scid mice 49 50, Dμ transcripts resulting from DH–JH rearrangement were readily detectable in scid and bcl-2 scid bone marrow.
Figure 1.
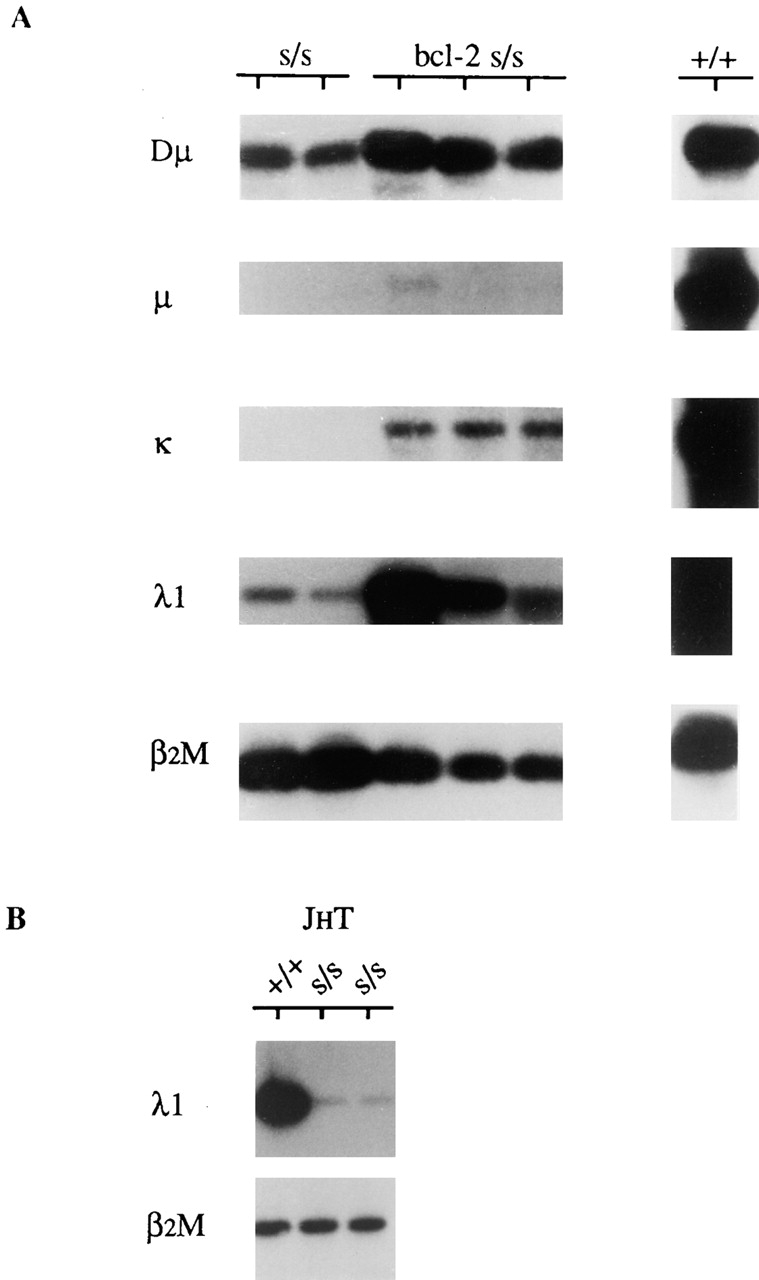
(A) Detection of λ1 transcripts in bone marrow of individual scid (s/s) and bcl-2 s/s mice and (B) in pooled bone marrow of JHT s/s and JHT non-scid (+/+) mice. Transcripts were detected by RT-PCR using locus-specific primers (see Materials and Methods). Transcripts corresponding to DH–JH, VH–DJH, Vκ–Jκ, and Vλ1–Jλ1 rearrangements are denoted Dμ, μ, κ, and λ1, respectively. Amplification of β2M transcripts served as a control for the amount of input cDNA. Results obtained with wt (+/+) bone marrow are provided for comparison.
As differentiation of scid B lineage cells does not generally progress beyond the pro-B cell stage, our detection of λ1 transcripts in scid mice suggests that Vλ1–Jλ1 rearrangement may occur before the pre-B cell stage. To test whether this is indeed true and whether the λ1 locus can rearrange independently of the H chain locus, we assayed for λ1 transcripts in bone marrow of JHT mice. In these mice, B cell differentiation is completely arrested at the pro-B cell stage as a result of gene-targeted inactivation of the JH locus 24. Fig. 1 B shows that λ1 transcripts were readily detectable in JHT bone marrow. The abundance of λ1 transcripts in JHT mice was ∼40-fold less than in wt mice but ∼25-fold greater than in JHT scid mice (Table ). We conclude that Vλ1–Jλ1 rearrangement can occur at the pro-B cell stage and independently of H chain gene rearrangement.
Table 1.
Level of λ1 Transcripts in scid Fetal Liver and JHT scid Bone Marrow Relative to wt Controls
| Genotype | Fetal Liver | Bone Marrow |
|---|---|---|
| +/+ | 1.0 | 1.0 |
| s/s | 1.2, 0.52 | – |
| (0.76, 0.83) | – | |
| JHT s/s | – | 0.001 |
| JHT +/+ | – | 0.024 |
PCR-amplified (23 cycles) λ1 transcripts, and also Vλ1–Jλ1 rearrangements in the case of fetal liver, were gel electrophoresed, blotted, and hybridized with a λ1-specific probe (see Materials and Methods). The values shown correspond to the amount of λ1 hybridizing signal normalized against the internal control (β2M) and the reference control, fetal liver or bone marrow of wt (+/+) mice. Thus, for example, the ratio of λ1/β2M hybridizing signal in JHT (JHT +/+) bone marrow divided by the λ1/β2M hybridizing signal in the reference control equaled 0.024. Two values are shown for scid fetal liver. These correspond to the ratios obtained for day 13 and 14 samples, respectively. The two values in parentheses correspond to the ratios obtained for Vλ1–Jλ1 rearrangement in genomic DNA from day 13 and 14 scid fetal liver, respectively.
Developmental Onset of Vλ1–Jλ1 Rearrangement in scid and wt Embryos.
To compare the developmental onset of Vλ1–Jλ1 rearrangement with that at other Ig loci, we tested genomic DNA from pooled livers of scid and wt embryos for DH–JH, VH–DJH, Vκ–Jκ, and Vλ1–Jλ1 rearrangement (Fig. 2). In wt embryos, we found that DH–JH rearrangement could be detected as early as day 12, whereas VH–DJH and Vκ–Jκ rearrangements were not detectable until day 14. These results are in general agreement with earlier reports on the time course of H and κ chain gene rearrangement in fetal mice 51 52. In scid embryos, DH–JH rearrangement was not clearly evident until day 14, and VH–DJH and Vκ–Jκ rearrangements were not detected except for a VH–DJH rearrangement of aberrant size in the day 15 sample. In contrast, Vλ1–Jλ1 rearrangement was evident as early as day 12 in both scid and wt embryos. These results indicate that Vλ1–Jλ1 rearrangement is initiated early in development and may developmentally precede VH–DJH and Vκ–Jκ rearrangement.
Figure 2.
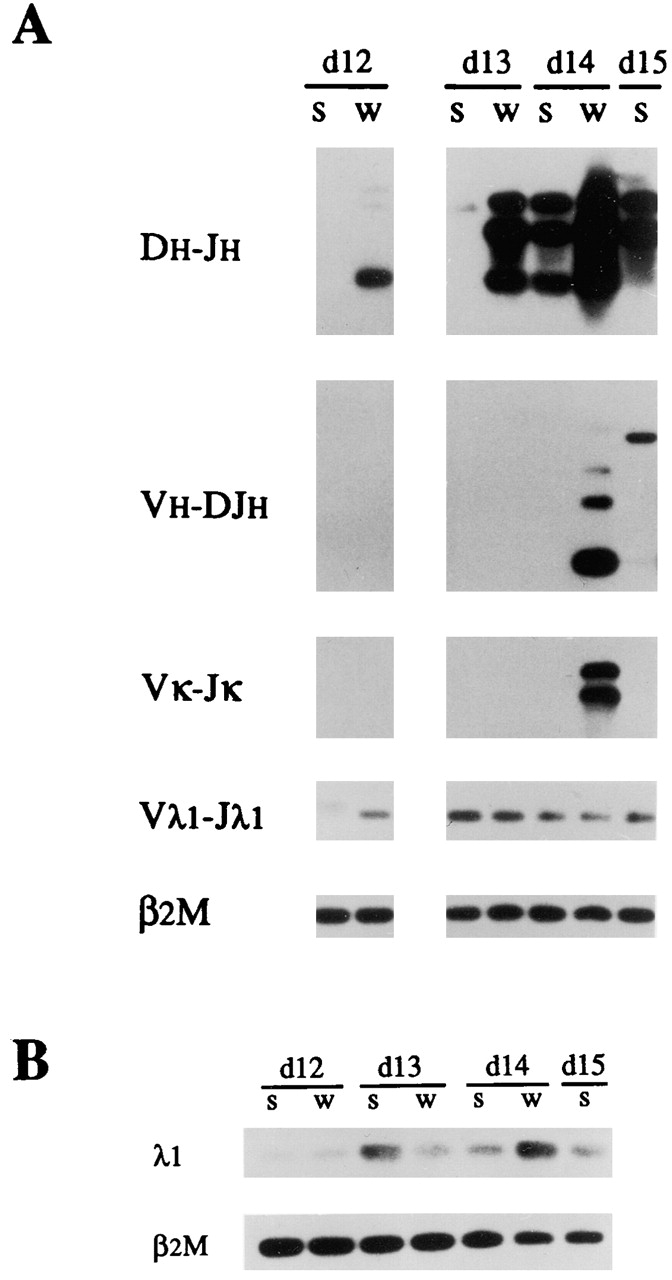
Developmental onset of Vλ1–Jλ1 rearrangement relative to that of DH–JH, VH–DJH, and Vκ–Jκ rearrangement in fetal liver of scid (s) and wt (w) mice. (A) Genomic DNA from pooled fetal liver at day (d) 12, 13, 14, and 15 of gestation was subjected to PCR using locus-specific primers to amplify DH–JH1-3, VH–DJH1-4, Vκ–Jκ1,2, and Vλ1–Jλ1 rearrangements. Amplification of the nonrearranging β2M gene served as a control for input DNA. (B) Amplification of λ1 and β2M transcripts was done by RT-PCR.
It is important to note in Fig. 2A and Fig. B, that the level of Vλ1–Jλ1 rearrangement and λ1 transcript in day 13–15 scid fetal liver remains relatively constant and appears comparable to that in the day 13 and 14 wt fetal liver. Indeed, quantitation of the amount of λ1 hybridizing signal for Vλ1–Jλ1 rearrangement and λ1 transcript in the day 13 and 14 scid samples showed this to be ∼80% of that in the corresponding wt samples (Table ). It should be noted that the observed level of Vλ1–Jλ1 rearrangement in DNA from 106 fetal liver cells was about two orders of magnitude less than in control DNA samples from 106 adult bone marrow cells of wt mice (data not shown). This is not surprising, as day 13–14 fetal liver lacks detectable pre-B cells and contains ≤1% pro-B cells (reference 53 and our unpublished results).
Vλ1–Jλ1 Rearrangements from scid Mice Contain Abnormal Deletions at the VJ Junction.
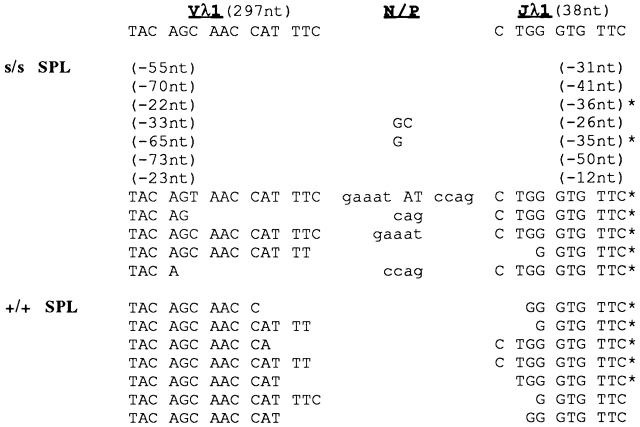
PCR-amplified Vλ1–Jλ1 rearrangements were detectable not only in scid fetal liver and adult bone marrow, but also in scid adult spleen. In most of these rearrangements, the Vλ1 and/or Jλ1 coding segments were abnormally truncated by >20 nucleotides. Deletions of this magnitude were not observed in Vλ1–Jλ1 rearrangements from wt mice. Representative results are illustrated in Fig. 3 and Fig. 4 for adult bone marrow and spleen, respectively. More than 70% of scid Vλ1–Jλ1 rearrangements (121/147 distinct sequences analyzed) contained abnormal deletions; those lacking such deletions often showed unusually long P additions, as illustrated in Fig. 4.
Figure 3.
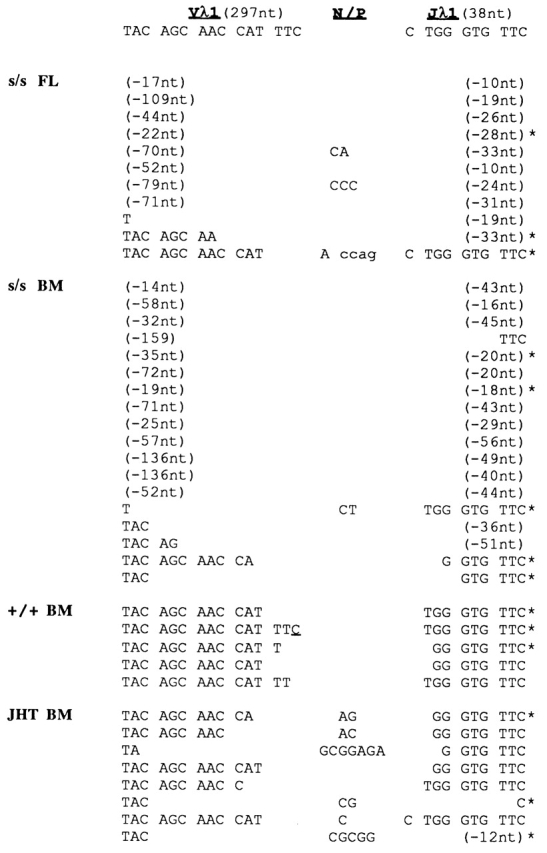
Representative Vλ1–Jλ1 junctional sequences in genomic DNA from pooled fetal liver (FL) of scid (s/s) embryos (day 13) and from pooled bone marrow (BM) of s/s, wt (+/+), and JHT adult mice. Germline nucleotides (nt) for the 3′ and 5′ ends of the Vλ1 and Jλ1 gene are shown at the top (the germline Vλ1 and Jλ1 coding regions comprise 297 and 38 nt, respectively). Upper- and lowercase letters under the N/P column denote N and P nucleotide additions, respectively. The number of V or J nucleotides deleted from the Vλ1 or Jλ1 coding end is indicated in parentheses; the asterisk denotes an in-frame rearrangement.
Figure 4.
Representative Vλ1–Jλ1 junctional sequences in genomic DNA of pooled spleen (SPL) from scid (s/s) and wt (+/+) adult mice. Format is as in Fig. 3.
Vλ1–Jλ1 rearrangements from bone marrow of JHT mice, in contrast to those from bone marrow of scid mice, showed nontemplated (N) additions and comparatively small deletions at the VJ junction (Fig. 3). N addition is dependent on terminal deoxynucleotidyl transferase (TdT) 54 55. This enzyme is expressed at the pro-B cell stage 56, the stage at which B cell differentiation is arrested in JHT mice 24. The absence of N additions in Vλ1–Jλ1 junctions from wt mice (Fig. 3 and Fig. 4) is in agreement with earlier reports 57 58 and consistent with the occurrence of most L chain rearrangement at the late pre-B cell stage 5 6, when TdT expression is dramatically downregulated 56. As scid and JHT mice both show an arrest of B cell differentiation at the pro-B stage, the abnormal loss of nucleotides in scid Vλ1–Jλ1 junctions must reflect the effect of the scid mutation and not a peculiarity of premature Vλ1–Jλ1 rearrangement.
λ1 Transcripts in scid Adult Bone Marrow and Spleen Lack Abnormal Deletions at Their VJ Junction.
Most λ1 transcripts from scid adult bone marrow and spleen corresponded to in-frame Vλ1–Jλ1 rearrangements with frequent P additions (illustrated in Fig. 5). The P additions consisted of three to five nucleotides (cag, ccag, and gaaat) and were found repeatedly in individual mice. The most common recurring sequence consisted of two P additions separated by an AT dinucleotide (gaaat-AT-ccag). Interestingly, the AT dinucleotide is palindromic to the last two nucleotides of the (gaaat) P addition. Similar restricted VJ junctional sequences and recurring P additions were also observed in λ1 transcripts recovered from bcl-2 scid spleen (data not shown). It is important to note that each PCR amplification of bone marrow and splenic cDNA from scid mice was done in parallel with PCR amplification of splenic cDNA from wt and RAG1−/− mice. We found that λ1 transcripts from three individual wt mice lacked N/P additions (illustrated in Fig. 5). No λ1 transcripts were recovered from cDNA of RAG1−/− mice. Thus, the observed recurring P additions appear to be a unique property of λ1 transcripts in the bone marrow and spleens of scid mice.
Figure 5.

Representative VJ junctional sequences in λ1 transcripts from individual scid (s/s), wt (+/+), and JHT scid mice. Both bone marrow (BM) and spleen (SPL) of three s/s and three +/+ mice was analyzed; the results for two individuals are shown. Format is as in Fig. 3.
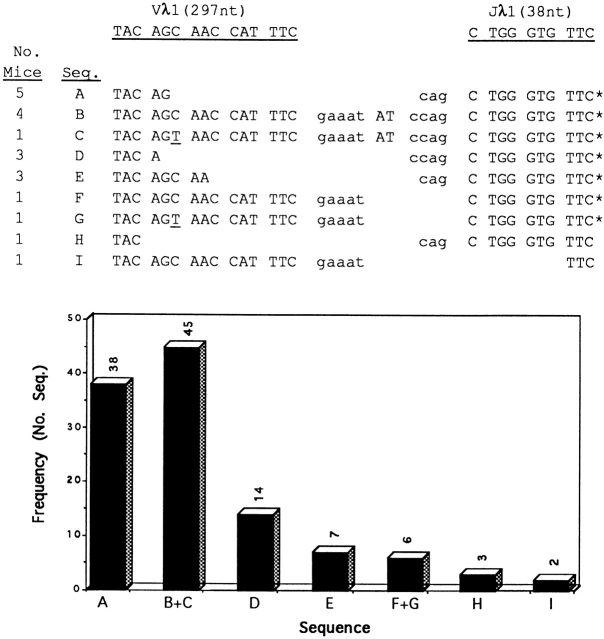
We analyzed a total of 188 cloned sequences from λ1 transcripts in bone marrow and/or spleens of five individual scid mice. We found that 115 clones contained P additions. In Fig. 6, each distinct junctional sequence (denoted A–I) among the 115 clones is listed according to its representation in individual mice and overall frequency (see histogram). Sequences B and C are treated as one in the histogram, as are sequences F and G, because each of these pairs is identical except for the substitution of T (underlined) for C in the Vλ1 germline codon, AGC. Note that (a) sequences A–G, comprising most of the clones (110/115), corresponded to in-frame Vλ1–Jλ1 rearrangements; (b) sequences A, B, and C accounted for ∼70% (83/115) of the Vλ1–Jλ1 rearrangements; and (c), the gaaat-AT-ccag P addition was present in all mice analyzed and represented nearly 40% of the clones (45/115).
Figure 6.
Recurring P additions in VJ junctions of λ1 transcripts from scid adult mice. Bone marrow and/or spleens of five individual scid mice were analyzed. We obtained VJ junctional sequences from 188 clones and found that 115 of these contained P additions. 9 distinct VJ junctional sequences were found among the 115 clones; these are denoted (A–I) below the underlined germline sequence for the Vλ1 and Jλ1 coding ends. The number of mice that contained a given VJ junctional sequence is indicated at left. The asterisk denotes that the rearrangements were in frame. The overall frequency of each distinct junctional sequence (A–I) is shown in the histogram.
In contrast to scid mice, λ1 transcripts from JHT scid mice lacked P additions and showed abnormal deletions at their VJ junctions similar to scid genomic Vλ1–Jλ1 rearrangements; furthermore, most corresponded to out-of-frame Vλ1–Jλ1 rearrangements (illustrated in Fig. 5). These results suggest, as discussed later, that survival of λ1-expressing scid cells could depend on the coexpression of a Dμ (or μ) chain. Consistent with this possibility, Dμ and λ1 transcripts were the only Ig gene transcripts routinely detectable in scid bone marrow (illustrated in Fig. 1). Moreover, most scid Dμ transcripts (10/16 analyzed) corresponded to DH–JH rearrangements in reading frame 2 (data not shown), which would be expected to result in the expression of a Dμ chain.
Cell Surface Expression of λ1 Chains Is Detectable in Spleens of bcl-2 scid Mice.
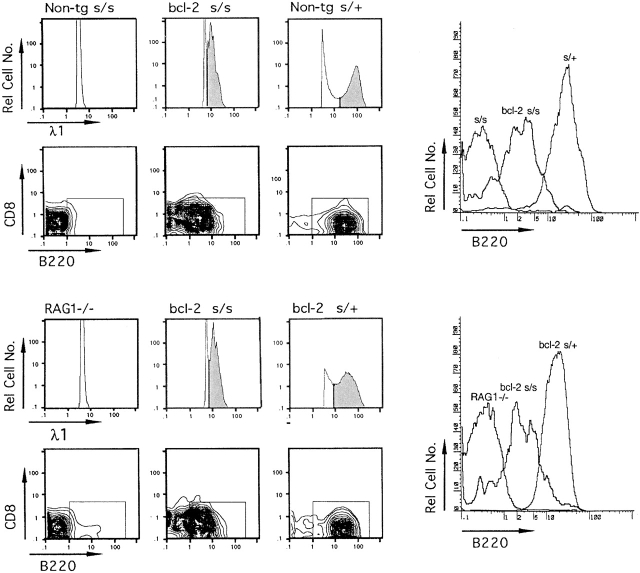
The presence of λ1-expressing scid cells in the spleen prompted us to test for possible cell surface expression of λ1 chains (λ1+ cells) in the spleens of scid and bcl-2 scid mice. The latter mice were included because survival of scid B lineage cells is known to be enhanced in the presence of the bcl-2 transgene 42. Large numbers (5–10 × 106) of cells were analyzed by three-color flow cytometry for expression of cell surface λ1 and the B and T cell markers CD45 (B220) and CD8, respectively. Mice with an inactivated RAG1 locus (RAG1−/− mice; reference 45) served as a negative control for background staining of λ1. As illustrated in Fig. 7, there were no detectable λ1+B220+CD8− cells in scid and RAG1−/− mice. However, in bcl-2 scid mice, λ1+B220+CD8− cells were detectable at a frequency of 0.02–0.05% versus 0.5–1.5% in the wt controls. The distribution of B220 staining for λ1 gated cells is shown in the histograms on the right side of Fig. 7. Note that bcl-2 scid spleen cells stained less bright for B220 and λ1 than scid/+ or bcl-2 scid/+ spleen cells. These results indicate that a low frequency of scid cells express λ1 chains on their cell surfaces and can be detected in the spleen of bcl-2 scid mice.
Figure 7.
Detection of λ1-expressing cells in spleens of bcl-2 scid (s/s) mice by three-color flow cytometry. Gate settings for scoring λ1+ cells in bcl-2 s/s mice were based on the distribution of λ1 staining in positive and negative controls, bcl-2 s/+ and RAG−/− mice, respectively. λ1+ cells in bcl-2 s/s and s/+ control mice are denoted in the shaded areas of the histograms for relative cell number versus λ1 staining. The λ1+ cells in the shaded areas were analyzed for CD8 and B220 expression; most of these cells (>85%) displayed a B cell phenotype (λ1+B220+CD8−) and fell within the boxed areas. The λ1+ cells in bcl-2 s/s mice represented ∼0.02–0.05% of the spleen cells analyzed and showed a λ1dullB220dull phenotype. In contrast, λ1+ cells in s/+ and bcl-2 s/+ mice were λ1brightB220bright and represented ∼0.5–1.5% of the spleen cells analyzed. The overlay of B220 histograms on the far right shows the distribution of B220 staining for cells in the s/s and RAG−/− mice and for λ1+ cells in the bcl-2 s/s, s/+, and bcl-2 s/+ mice.
Three-color flow cytometry was used also to analyze bcl-2 scid mice for expression of surface μ chains (μ+ cells). No μ+ cells were detected (data not shown), consistent with previous reports showing a lack of μ+ cells in bcl-2 scid spleen 42 59.
Discussion
The preceding results support earlier evidence, cited in the Introduction, that initiation of rearrangement at the λ locus does not require prior rearrangement at the H or κ chain locus. Our detection of λ1 transcripts in bone marrow of scid and JHT mice indicates that Vλ1–Jλ1 rearrangement can occur before the pre-B cell stage and independently of H chain gene rearrangement. Moreover, the detection of Vλ1–Jλ1 rearrangement as early as day 12 in wt fetal liver, in which there is no genetic impairment of Ig gene rearrangement, suggests that the onset of Vλ1–Jλ1 rearrangement may developmentally precede VH–DJH and Vκ–Jκ rearrangement. The latter rearrangements were not detectable before day 14.
The most novel aspect of our findings is the regular detection of Vλ1–Jλ1 rearrangement in scid mice. This would not have been predicted, particularly the comparable level of Vλ1–Jλ1 rearrangement in scid and wt fetal liver. Also unexpected are the recurring P additions in scid λ1 transcripts and the presence of scid cells with in-frame Vλ1–Jλ1 rearrangements in the spleen. We discuss the implications of these findings below.
Levels of Vλ1–Jλ1 Rearrangement and λ1 Transcript in scid Mice.
We were surprised to find that the level of Vλ1–Jλ1 rearrangement in scid fetal liver was comparable to that in wt fetal liver (Table ), despite the abnormal loss of nucleotides in the VJ junction of most scid Vλ1–Jλ1 rearrangements. Thus, in scid fetal liver, abnormally truncated Vλ1 and Jλ1 coding ends appear to be joined as efficiently as Vλ1 and Jλ1 coding ends in wt fetal liver. This is unexpected because previous studies have clearly shown that V(D)J rearrangement is severely impaired in B and T lineage cells of adult scid mice 31 32 33 34. Indeed, as discussed below, Vλ1–Jλ1 rearrangement in scid adult bone marrow cells appears to be much less efficient than in wt bone marrow cells. The basis for the comparable level of Vλ1–Jλ1 rearrangement in scid and wt fetal liver is unexplained. Possibly, the machinery available for nonhomologous end joining in fetal liver differs from that in adult bone marrow and is able to compensate for the scid deficiency in DNA-PKcs.
Whereas the level of λ1 transcripts was comparable in scid and wt fetal liver, in scid adult bone marrow (from JHT scid mice), λ1 transcript levels were ∼25-fold less than in control adult bone marrow of JHT mice (Table ). The latter comparison is valid because B cell differentiation is arrested at the same stage (pro-B cell stage) in both JHT and JHT scid mice. Moreover, neither JHT nor JHT scid mice can make Dμ or μ chains, which could potentially affect the selection (or survival) of cells with productive λ1 transcripts.
Two explanations can be considered for the 25-fold difference in levels of λ1 transcript in JHT and JHT scid adult bone marrow. The first postulates that the scid λ1 transcripts are much less stable than those generated in wt bone marrow, possibly owing to the abnormal nucleotide deletions accompanying scid Vλ1–Jλ1 rearrangement. We would expect such instability to be manifest in scid fetal liver as well, and yet in this tissue the level of λ1 transcript was comparable to that in wt fetal liver. A second explanation is that the 25-fold difference in abundance of λ1 transcripts in JHT and JHT scid mice primarily reflects a lower efficiency of Vλ1–Jλ1 rearrangement in scid adult pro-B cells than wt adult pro-B cells. This explanation is consistent with previous studies showing a 10–20-fold lower level of DH–JH rearrangement in scid than in wt adult bone marrow 49 50.
The question arises as to why we did not obtain evidence of premature Vκ–Jκ rearrangement in scid bone marrow, given that a low level of such rearrangement has been previously observed in the pro-B cell fraction of wt mice 5 6. One possible reason is that initiation of Vκ–Jκ rearrangement invariably results in cell death. Consistent with this possibility, κ transcripts were readily detectable in the bone marrow of bcl-2 scid mice. The bcl-2 transgene is known to promote the longevity of scid B lineage cells 42 and their differentiation beyond the pro-B cell stage 42 59. As reasoned elsewhere 60, primary and secondary initiation of Vκ–Jκ rearrangement in scid cells lacking the bcl-2 transgene would be expected to result in persisting chromosomal breaks and cell death.
VJ Junctions of scid λ1 Transcripts Show Recurring P Additions.
Most λ1 transcripts recovered from scid adult mice corresponded to in-frame Vλ1–Jλ1 rearrangements, and >60% of these transcripts contained recurring P additions. Whereas in wt mice most P additions are one or two nucleotides 61, in scid mice they are often longer 62 63. In the present case, all of the scid P additions at Vλ1 were five nucleotides in length (gaaat), and those at Jλ1 were either three or four nucleotides in length (cag or ccag). This may reflect a strong bias in the resolution of hairpin coding ends imposed by the scid DNA-PKcs deficiency, such that Jλ1 and Vλ1 coding ends are frequently nicked three to four and five nucleotides from the hairpin tip, respectively, and then joined without further modification. In addition, cells expressing λ1 transcripts with these junctions may be strongly selected, as discussed later.
Of particular interest is the recurring Vλ1–Jλ1 junctional sequence of two P additions separated by an AT dinucleotide (gaaat-AT-ccag), which was present in ∼40% of the λ1 transcripts with P additions (Fig. 6). The basis for the AT dinucleotide is unclear. Given that scid Vλ1–Jλ1 rearrangements occur at the pro-B cell stage in the presence of high TdT levels, the AT dinucleotide could represent a nontemplated addition mediated by TdT. Another possibility is that the AT dinucleotide, which is palindromic to the last two nucleotides of the gaaat P addition, corresponds to a secondary P addition. In this scenario, one could postulate two successive recombination events. The generation of the gaaat P addition would result from an open and shut recombination 64 at Vλ1. This would be followed by secondary cleavage at the Vλ1 signal/coding border, asymmetric nicking of the Vλ1 hairpin coding end, and joining to a Jλ1 coding end with a ccag overhang. Regardless of how the AT dinucleotide is generated, we suggest that cells with the gaaat-AT-ccag junctional sequence are strongly selected to account for the repeated occurrence of this sequence in λ1 transcripts of individual scid mice.
Evidence for λ1-Expressing scid Cells.
Scid λ1 transcripts were not only detected in bone marrow but also in spleen. Moreover, λ1 transcripts with the same VJ junctional sequences were often found to recur in both of these tissues. This implies a strong selection for cells with in-frame Vλ1–Jλ1 rearrangements containing particular VJ junctional sequences. Such selection was not evident in JHT scid mice. λ1 transcripts from JHT scid bone marrow contained abnormal deletions at the VJ junction, lacked P additions, and in most cases corresponded to out-of-frame Vλ1–Jλ1 rearrangements. These findings suggest that survival and selection of λ1-expressing cells requires a functional H chain locus.
How might λ1-expressing scid cells be dependent on a functional H chain locus? One possibility is that Dμ and λ1 chains, resulting from expression of in-frame Dμ and λ1 transcripts in scid bone marrow, pair and associate with the Ig α and β signal-transducing chains 65 66 to form a B cell–like receptor (BCRDμ/λ1). Although Dμ chains cannot pair efficiently with κ chains 67, they can pair with the surrogate L chain 68 69 and might be expected to pair with λ1 chains as well because the latter share some homology with the surrogate L chain (for review see reference 70). Expression of BCRDμ/λ1 in this proposed scenario would signal rapid (or direct) progression of scid pro-B cells to the recombinase-inactive B cell stage and allow these cells to survive and migrate to the periphery. But as BCRDμ/λ1 would lack a VH region, we would not expect cells bearing this receptor to persist or expand in response to naturally occurring antigens. This could in part explain the very low frequency of cells with surface λ1 in bcl-2 scid spleen (≤0.05% of the cells examined). What is experimentally missing in support of the above scenario, however, is evidence for bcl-2 scid cells with surface λ1 and μ chains. Despite reported evidence for intracellular expression of μ (or Dμ) chains in B220+CD22+ spleen cells of bcl-2 scid mice 59, cells with surface μ chains have not been detected in bcl-2 scid mice 42 59.
In conclusion, we suggest that (a) joining of Vλ1 and Jλ1 coding ends after the initiation of Vλ1–Jλ1 rearrangement may not be impaired in B lineage cells of scid fetal liver because of the developmental time at which such rearrangement occurs; (b) recurring P additions in scid λ1 transcripts may reflect a strong bias in the resolution of Vλ1 and Jλ1 hairpin coding ends imposed by the scid defect as well as possible strong selection for cells expressing these λ1 transcripts; and (c) pro-B cells with in-frame Vλ1–Jλ1 rearrangements may express a pre-BCR–like receptor and differentiate into recombinase-inactive cells and emigrate from bone marrow to spleen.
Acknowledgments
We thank Kerry Campbell, Randy Hardy, Dietmar Kappes, Pam Nakajima, Gillian Wu, and Martin Weigert for review of the manuscript and Samuel Litwin for helpful discussion. We also thank Roseanne Diehl for assistance in typing the manuscript.
Grants from the National Institutes of Health (CA06927 and CA04946) and an appropriation from the Commonwealth of Pennsylvania supported this work.
Footnotes
Abbreviations used in this paper: DNA-PKcs, DNA protein kinase catalytic subunit; JHT, JH targeted deletion; P, palindromic; RAG, recombination activation gene; RT, reverse transcriptase; TdT, terminal deoxynucleotidyl transferase; wt, wild-type.
References
- Yancopoulos G.D., Alt F.W. Regulation of the assembly and expression of variable-region genes. Annu. Rev. Immunol. 1986;4:339–368. doi: 10.1146/annurev.iy.04.040186.002011. [DOI] [PubMed] [Google Scholar]
- Gorman J.R., Alt F.W. Regulation of immunoglobulin light chain isotype expression. Adv. Immunol. 1998;69:113–181. doi: 10.1016/s0065-2776(08)60607-0. [DOI] [PubMed] [Google Scholar]
- Alt F.W., Yancopoulos G.D., Blackwell T.K., Wood C., Thomas E., Boss M., Coffman R., Rosenberg N., Tonegawa S., Baltimore D. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO (Eur. Mol. Biol. Organ.) J. 1984;3:1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy R.R., Carmack C.E., Shinton S.A., Kemp J.D., Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J. Exp. Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlich A., Schaal S., Gu H., Kitamura D., Muller W., Rajewsky K. Immunoglobulin heavy and light chain genes rearrange independently at early stages of B cell development. Cell. 1993;72:695–704. doi: 10.1016/0092-8674(93)90398-a. [DOI] [PubMed] [Google Scholar]
- Li Y.-S., Hayakawa K., Hardy R.R. The regulated expression of B lineage associated genes during B cell differentiation in bone marrow and fetal liver. J. Exp. Med. 1993;178:951–960. doi: 10.1084/jem.178.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt F.W., Enea V., Bothwell A.L.M., Baltimore D. Activity of multiple light chain genes in murine myeloma cells producing a single functional light chain. Cell. 1980;21:1–12. doi: 10.1016/0092-8674(80)90109-9. [DOI] [PubMed] [Google Scholar]
- Hieter P., Korsmeyer S., Waldman T., Leder P. Human immunoglobulin κ light-chain genes are deleted or rearranged in λ-producing cells. Nature. 1981;290:368–372. doi: 10.1038/290368a0. [DOI] [PubMed] [Google Scholar]
- Korsmeyer S.I., Hieter P.A., Sharrow S.O., Goldman C.K., Leder P., Waldman T.A. Normal human B cells display ordered light chain gene rearrangement and deletions. J. Exp. Med. 1982;156:975–985. doi: 10.1084/jem.156.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel H., Rolink A., Weiss S. B cells are programmed to activate κ and λ for rearrangement at consecutive developmental stages. Eur. J. Immunol. 1999;29:2167–2176. doi: 10.1002/(SICI)1521-4141(199907)29:07<2167::AID-IMMU2167>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Coleclough C., Perry R.P., Karjalainen K., Weigert M. Aberrant rearrangements contribute significantly to the allelic exclusion of immunoglobulin gene expression. Nature. 1981;290:372–378. doi: 10.1038/290372a0. [DOI] [PubMed] [Google Scholar]
- Ramsden D.A., Wu G.E. Mouse κ light-chain recombination signal sequences mediate recombination more frequently than do those of λ light chain. Proc. Natl. Acad. Sci. USA. 1991;88:10721–10725. doi: 10.1073/pnas.88.23.10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Jenab J., Rosenberg N. κ and λ rearrangement occur simultaneously in transformed pre-B cells. J. Immunol. 1997;159:6061–6069. [PubMed] [Google Scholar]
- Kubagawa H., Cooper M.D., Carroll A.J., Burrows P.D. Light-chain gene expression before heavy-chain gene rearrangement in pre-B cells transformed by Epstein-Barr virus. Proc. Natl. Acad. Sci. USA. 1989;86:2356–2360. doi: 10.1073/pnas.86.7.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlissel M.S., Baltimore D. Activation of immunoglobulin kappa gene rearrangement correlates with induction of germline kappa gene transcription. Cell. 1989;58:1001–1007. doi: 10.1016/0092-8674(89)90951-3. [DOI] [PubMed] [Google Scholar]
- Felsher D.W., Ando D.T., Braun J. Independent rearrangement of Igλ genes in tissue culture-derived murine B cell lines. Int. Immunol. 1991;3:711–718. doi: 10.1093/intimm/3.7.711. [DOI] [PubMed] [Google Scholar]
- Chen J., Trounstine M., Kurahara C., Young F., Kuo C.-C., Xu Y., Loring J.F., Alt F.W., Huszar D. B cell development in mice that lack one or both immunoglobulin κ light chain genes. EMBO (Eur. Mol. Biol. Organ.) J. 1993;12:821–830. doi: 10.1002/j.1460-2075.1993.tb05722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y.-R., Takeda S., Rajewsky K. Gene targeting in the Igκ locusefficient generation of chain-expressing B cells, independent of gene rearrangements in Igκ. EMBO (Eur. Mol. Biol. Organ.) J. 1993;12:811–820. doi: 10.1002/j.1460-2075.1993.tb05721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy R.R., Dangl J.L., Hayakawa K., Jager G., Herzenberg L., Herzenberg L.A. Frequent lambda light chain gene rearrangement and expression in a Ly-1 B lymphoma with a productive kappa chain allele. Proc. Natl. Acad. Sci. USA. 1986;83:1438–1442. doi: 10.1073/pnas.83.5.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg J., McDowell M., Jack H.-M., Wabl M. Immunoglobulin λ gene rearrangement can precede κ gene rearrangement. Dev. Immunol. 1990;1:53–57. doi: 10.1155/1990/56014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J.-Q., Bene M.C., Faure G.C. Alternative rearrangements of immunoglobulin light chain genes in human leukemia. Leukemia. 1991;5:651–656. [PubMed] [Google Scholar]
- Gollahon K.A., Hagman J., Brinster R.L., Storb U. Ig λ-producing B cells do not show feedback inhibition of gene rearrangement. J. Immunol. 1988;141:2771–2780. [PubMed] [Google Scholar]
- Coleclough C. Chance, necessity and antibody gene dynamics. Nature. 1983;303:23–26. doi: 10.1038/303023a0. [DOI] [PubMed] [Google Scholar]
- Chen J., Trounstine M., Alt F.W., Young F., Kurahara C., Loring J.F., Huszar D. Immunoglobulin gene rearrangement in B cell deficient mice generated by targeted deletion of the JH locus. Int. Immunol. 1993;5:647–656. doi: 10.1093/intimm/5.6.647. [DOI] [PubMed] [Google Scholar]
- Fulop G.M., Phillips R.A. The scid mutation in mice causes a general defect in DNA repair. Nature. 1990;347:479–482. doi: 10.1038/347479a0. [DOI] [PubMed] [Google Scholar]
- Hendrickson E.A., Qin X.-Q., Bump E.A., Schatz D.G., Oettinger M., Weaver D.T. A link between double-strand break-related and V(D)J recombinationthe scid mutation. Proc. Natl. Acad. Sci. USA. 1991;88:4061–4065. doi: 10.1073/pnas.88.10.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedermann K.A., Sun J., Giaccia A.J., Tosto L.M., Brown M. Scid mutation in mice confers hypersensitivity to ionizing radiation and a deficiency in DNA double-strand break repair. Proc. Natl. Acad. Sci. USA. 1991;88:1394–1397. doi: 10.1073/pnas.88.4.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchgessner C.U., Patil C.K., Evans J.W., Cuomo C.A., Fried L.M., Carter T., Oettinger M., Brown M.J. DNA-dependent kinase (p350) as a candidate gene for the murine SCID defect. Science. 1995;267:1178–1182. doi: 10.1126/science.7855601. [DOI] [PubMed] [Google Scholar]
- Blunt T., Gell D., Fox M., Taccioli G.E., Lehmann A.R., Jackson S.P., Jeggo P.A. Identification of a nonsense mutation in the carboxyl-terminal region of DNA-dependent protein kinase catalytic subunit in the scid mouse. Proc. Natl. Acad. Sci. USA. 1996;93:10285–10290. doi: 10.1073/pnas.93.19.10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danska J., Holland D.P., Mariathasan S., Williams K.M., Guidos C.J. Biochemical and genetic defects in the DNA-dependent protein kinase in murine scid lymphocytes. Mol. Cell. Biol. 1996;16:5507–5517. doi: 10.1128/mcb.16.10.5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler W., Weiler I.J., Schuler A., Phillips R.A., Rosenberg N., Mak T.W., Kearney J.F., Perry R.P., Bosma M.J. Rearrangement of antigen receptor genes is defective in mice with severe combined immune deficiency. Cell. 1986;46:963–972. doi: 10.1016/0092-8674(86)90695-1. [DOI] [PubMed] [Google Scholar]
- Lieber M.R., Hesse J.E., Lewis S., Bosma G.C., Bosma M.J., Gellert M. The defect in murine severe combined immune deficiencyjoining of signal segments but not coding segments in V(D)J recombination. Cell. 1988;55:7–16. doi: 10.1016/0092-8674(88)90004-9. [DOI] [PubMed] [Google Scholar]
- Malynn B.A., Blackwell T.K., Fulop G.M., Rathburn G.A., Furley A.J.W., Ferrier P., Heinke L.B., Phillips R.A., Yancopoulos G.D., Alt F.W. The scid defect affects the final step of the immunoglobulin VDJ recombinase mechanism. Cell. 1988;54:453–460. doi: 10.1016/0092-8674(88)90066-9. [DOI] [PubMed] [Google Scholar]
- Blackwell T.K., Malynn B.A., Pollock R.R., Ferrier P., Covey L.R., Fulop G.M., Phillips R.A., Yancopoulos G.D., Alt F.W. Isolation of scid pre-B cells that rearrange kappa light chain genesformation of normal signal and abnormal coding joins. EMBO (Eur. Mol. Biol. Organ.) J. 1989;8:735–742. doi: 10.1002/j.1460-2075.1989.tb03433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack W.T., Tjoelker L.W., Carlson L.M., Petryniak B., Barth C.F., Humphries E.H., Thompson C.B. Chicken IgL gene rearrangement involves deletion of a circular episome and addition of single nonrandom nucleotides to both coding segments. Cell. 1989;56:785–791. doi: 10.1016/0092-8674(89)90683-1. [DOI] [PubMed] [Google Scholar]
- Lafaille J.J., De Cloux A., Bonneville M., Takagaki Y., Tonegawa S. Junctional sequences of T cell receptor γδ genesimplications for γδ T cell lineages and for a novel intermediate of V-D-J joining. Cell. 1989;59:859–870. doi: 10.1016/0092-8674(89)90609-0. [DOI] [PubMed] [Google Scholar]
- Lieber M.R. Site-specific recombination in the immune system. FASEB J. 1991;5:2934–2944. doi: 10.1096/fasebj.5.14.1752360. [DOI] [PubMed] [Google Scholar]
- Roth D.B., Menetski J.P., Nakajima P.B., Bosma M.J., Gellert M. V(D)J recombinationbroken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell. 1992;70:983–991. doi: 10.1016/0092-8674(92)90248-b. [DOI] [PubMed] [Google Scholar]
- van Gent D.C., McBlane J.F., Ramsden D.A., Sadofsky M.J., Hesse J.E., Gellert M. Initiation of V(D)J recombination in a cell-free system. Cell. 1995;81:925–934. doi: 10.1016/0092-8674(95)90012-8. [DOI] [PubMed] [Google Scholar]
- Ramsden D.A., Gellert M. Formation and resolution of double-strand break intermediates in V(D)J rearrangement. Genes Dev. 1995;9:2409–2420. doi: 10.1101/gad.9.19.2409. [DOI] [PubMed] [Google Scholar]
- Bosma G.C., Custer R.P., Bosma M.J. A severe combined immunodeficiency mutation in the mouse. Nature. 1983;30:527–530. doi: 10.1038/301527a0. [DOI] [PubMed] [Google Scholar]
- Strasser A., Harris A.W., Corcoran L.M., Cory S. Bcl-2 expression promotes B- but not T-lymphoid development in scid mice. Nature. 1994;368:457–460. doi: 10.1038/368457a0. [DOI] [PubMed] [Google Scholar]
- Drews R., Drohan W.N., Lubon H. Transgene detection in mouse tail digests. Biotechniques. 1994;17:866–867. [PubMed] [Google Scholar]
- Strasser A., Harris A.W., Huang D.C.S., Krammer P.H., Cory S. Bcl-2 and Fas/APO-1 regulate distinct pathways to lymphocyte apoptosis. EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:6136–6147. doi: 10.1002/j.1460-2075.1995.tb00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P., Iacomini J., Johnson R.S., Herrup K., Tonegawa S., Papaioannou V.E. RAG-1 deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- Chang Y., Bosma G.C., Bosma M.J. Development of B cells in scid mice with immunoglobulin transgenesimplications for the control of V(D)J recombination. Immunity. 1995;2:607–616. doi: 10.1016/1074-7613(95)90005-5. [DOI] [PubMed] [Google Scholar]
- Sakano H., Kurosawa Y., Weigert M., Tonegawa S. Identification and nucleotide sequence of a diversity DNA segment (D) of immunoglobulin heavy chain genes. Nature. 1981;290:562–565. doi: 10.1038/290562a0. [DOI] [PubMed] [Google Scholar]
- Marcu K.B., Banerji J., Penncavage N.A., Lang R., Arnheim N. 5′ flanking region of immunoglobulin heavy chain constant region genes displays length heterogeneity in germlines of inbred mouse strains. Cell. 1980;22:187–196. doi: 10.1016/0092-8674(80)90167-1. [DOI] [PubMed] [Google Scholar]
- Pennycook J.L., Chang Y., Celler J., Phillips R.A., Wu G.E. High frequency of normal DJH joints in B cell progenitors in severe combined immunodeficiency mice. J. Exp. Med. 1993;178:1007–1016. doi: 10.1084/jem.178.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki R., Itoh M., Hamatani K., Abe M. Normal D-JH rearranged products of the Ig H gene in SCID mouse bone marrow. Int. Immunol. 1996;8:1045–1053. doi: 10.1093/intimm/8.7.1045. [DOI] [PubMed] [Google Scholar]
- Chang Y., Paige C.J., Wu G.E. Enumeration and characterization of DJH structures in mouse fetal liver. EMBO (Eur. Mol. Biol. Organ.) J. 1992;11:1891–1899. doi: 10.1002/j.1460-2075.1992.tb05241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlissel M.S., Morrow T. Ig heavy chain protein controls B cell development by regulating gem-line transcription and retargeting V(D)J recombination. J. Immunol. 1994;153:1645–1657. [PubMed] [Google Scholar]
- Bosma G.C., Chang Y., Karasuyama H., Bosma M.J. Differential effect of an Ig μ transgene on development of pre-B cells in fetal and adult scid mice. Proc. Natl. Acad. Sci. USA. 1999;96:11952–11957. doi: 10.1073/pnas.96.21.11952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfillan S., Dierich A., Lemeur M., Benoist C., Mathis D. Mice lacking TdTmature animals with an immature lymphocyte repertoire. Science. 1993;261:1175–1178. doi: 10.1126/science.8356452. [DOI] [PubMed] [Google Scholar]
- Komori T., Okada A., Stewart V., Alt F.W. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science. 1993;261:1171–1175. doi: 10.1126/science.8356451. [DOI] [PubMed] [Google Scholar]
- Wasserman R., Li Y.-S., Hardy R.R. Down-regulation of terminal deoxynucleotidyl transferase by Ig heavy chain in B lineage cells. J. Immunol. 1997;158:1133–1138. [PubMed] [Google Scholar]
- Boudinot P., Drapier A.M., Cazenave P.A., Sanchez P. Mechanistic and selective constraints act on the establishment of V lambda J lambda junctions in the B cell repertoire. J. Immunol. 1994;152:2248–2255. [PubMed] [Google Scholar]
- Boudinot P., Rueff-Juy D., Drapier A.M., Cazenave P.A., Sanchez P. Various V-J rearrangement efficiencies shape the mouse lambda B cell repertoire. Eur. J. Immunol. 1995;25:2499–2505. doi: 10.1002/eji.1830250914. [DOI] [PubMed] [Google Scholar]
- Young F., Mizoguchi E., Bhan A.K., Alt F.W. Constitutive bcl-2 expression during immunoglobulin heavy chain-promoted B cell differentiation expands novel precursor B cells. Immunity. 1997;6:23–33. doi: 10.1016/s1074-7613(00)80239-3. [DOI] [PubMed] [Google Scholar]
- Chang Y., Bosma M.J. Effect of different Ig transgenes on B cell differentiation in scid mice. Int. Immunol. 1997;9:373–380. doi: 10.1093/intimm/9.3.373. [DOI] [PubMed] [Google Scholar]
- Meier J.T., Lewis S.M. P nucleotides in V(D)J recombinationa fine-structure analysis. Mol. Cell. Biol. 1993;13:1078–1092. doi: 10.1128/mcb.13.2.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler W., Ruetsch N.R., Amsler M., Bosma M.J. Coding joint formation of endogenous T cell receptor genes in lymphoid cells from scid miceunusual P-nucleotide additions in VJ-coding joints. Eur. J. Immunol. 1991;21:589–596. doi: 10.1002/eji.1830210309. [DOI] [PubMed] [Google Scholar]
- Kienker L.J., Kuziel W.A., Tucker P.W. T cell receptor γ and δ gene junctional sequences in SCID miceexcessive P nucleotide insertion. J. Exp. Med. 1991;174:769–773. doi: 10.1084/jem.174.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S.M., Hesse J.E. Cutting and closing without recombination in V(D)J joining. EMBO (Eur. Mol. Biol. Organ.) J. 1991;10:3631–3639. doi: 10.1002/j.1460-2075.1991.tb04929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reth M. Antigen receptors on B lymphocytes. Annu. Rev. Immunol. 1992;10:97–121. doi: 10.1146/annurev.iy.10.040192.000525. [DOI] [PubMed] [Google Scholar]
- Cambier J.C., Pleiman C.M., Clark M.R. Signal transduction by the B cell antigen receptor and its coreceptors. Annu. Rev. Immunol. 1994;12:457–486. doi: 10.1146/annurev.iy.12.040194.002325. [DOI] [PubMed] [Google Scholar]
- Horne M.C., Roth P.E., DeFranco A.L. Assembly of truncated immunoglobulin heavy chain Dμ into antigen receptor-like complexes in pre-B cells but not in B cells. Immunity. 1996;4:145–158. doi: 10.1016/s1074-7613(00)80679-2. [DOI] [PubMed] [Google Scholar]
- Tsubata T., Tsubata R., Reth M. Cell surface expression of the short immunoglobulin μ chain (Dμ protein) in murine pre-B cells is differently regulated from that of the intact μ chain. Eur. J. Immunol. 1991;21:1359–1363. doi: 10.1002/eji.1830210605. [DOI] [PubMed] [Google Scholar]
- Karasuyama H., Rolink A., Melchers F. A complex of glycoproteins is associated with VpreB/λ5 surrogate light chain on the surface of μ heavy chain–negative early precursor B cell lines. J. Exp. Med. 1993;178:469–478. doi: 10.1084/jem.178.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchers F., Karasuyama H., Haasner D., Bauer S., Kudo A., Sakaguchi N., Jameson B., Rolink A. The surrogate light chain in B-cell development. Immunol. Today. 1993;14:60–68. doi: 10.1016/0167-5699(93)90060-X. [DOI] [PubMed] [Google Scholar]