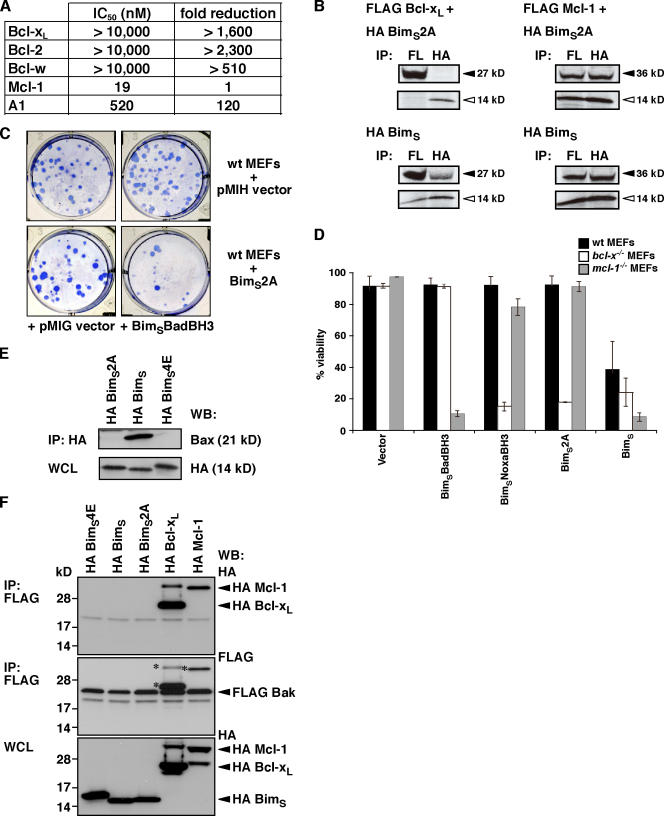
Figure 2.
Characterization of a novel Mcl-1–specific Bim variant. (A) A BimBH3 mutant that retains significant binding only to Mcl-1. The relative affinities of wild-type or the BimBH3 mutant 2A (L62A/F69A) peptides for prosurvival proteins were determined in solution competition assays using an optical biosensor. Compared with the wild-type sequence, the mutant 2A has reduced affinity for all prosurvival proteins except for Mcl-1. (B) Full-length BimS2A (top) interacts with Mcl-1 in cells (right), but, unlike wild-type BimS (bottom), it does not bind Bcl-xL (left), which is consistent with the in vitro binding assays using purified components (A). Prosurvival proteins are indicated by black arrowheads, and BimS/BimS2A is indicated by white arrowheads. (C) Functional cooperation between the Mcl-1–specific BimS2A variant and Bad. Stable pools of wild-type MEFs were generated by hygromycin selection after retroviral infection with the parental pMIH vector or one expressing the full-length Mcl-1–specific BimS2A mutant. Long-term colony formation was assessed when these pools were reinfected with the control pMIG retrovirus or one expressing BimSBadBH3 (a BimS variant with its BH3 replaced with that of Bad; Chen et al., 2005). BimS2A was inert on its own, but there was a striking reduction in colonies formed if BimSBadBH3, which targets the prosurvival proteins (Bcl-xL, Bcl-2, and Bcl-w) that BimS2A does not bind, is coexpressed. (D) The Mcl-1–specific BH3 ligands, full-length BimSNoxaBH3 or BimS2A, potently kill only when Bcl-xL is absent, whereas BimSBadBH3 only kills the cells deficient in Mcl-1. Cell viability was assessed 24 h after infection with the indicated retroviruses. (E) HA-tagged wild-type BimS but neither the BimS2A nor the inert BimS mutant (BimS4E) coimmunoprecipitated with endogenous Bax. (F) FLAG-tagged Bak (middle) coimmunoprecipitates with HA-tagged Bcl-xL and Mcl-1 but not with HA-tagged BimS4E, BimS, or BimS2A (top). Asterisks are the remnant signals from reprobing of the FLAG immunoprecipitation (top) with the FLAG antibody. Data in D represent means ± SD (error bars) of at least three experiments; those in A–C, E, and F) are from representative experiments. WCL, whole cell lysate.