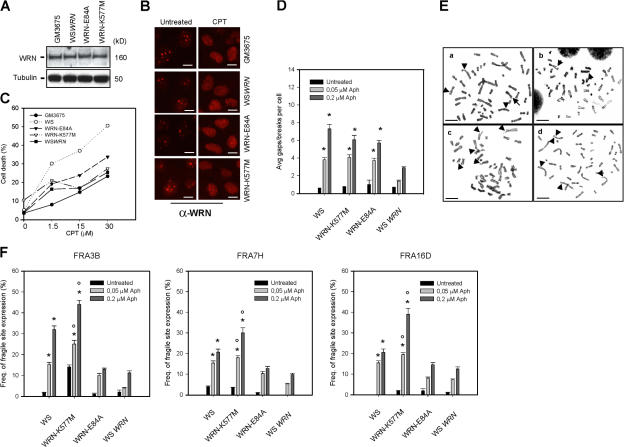
Figure 4.
Impaired WRN helicase activity is responsible for common fragile site instability. (A) Western blotting analysis showing the expression of WRN protein in cells stably expressing wild-type WRN (WSWRN) or missense mutant forms of WRN with impaired exonuclease (WRN-E84A) or helicase (WRN-K577M) activity. GM3675 fibroblasts were used as a positive control. The membrane was probed with α-WRN. Tubulin was used as a loading control. (B) Subnuclear localization of WRN in response to camptothecin-induced replication stress. Indirect immunofluorescence staining was achieved using the same antibody as in Western blotting. (C) Sensitivity of cells to camptothecin-induced replication stress. Cell death was evaluated by the trypan blue method as described in Cell death evaluation. The percentage of cell death was indicated for each dose of camptothecin. (D) Mean overall chromosome gaps and breaks per cell in WS cells, WS cells expressing mutant forms of exonuclease (WRN-E84A) or helicase (WRN-K577M) activity, and WS cells in which wild-type WRN was reintroduced (WSWRN). Cells were exposed to different doses of aphidicolin (Aph) 24 h before harvest. Data are presented as means of three independent experiments. (E) Representative Giemsa-stained metaphases of WS fibroblasts (a), WS cells transfected with wild-type WRN cDNA (WSWRN; b), or WRN lacking helicase (WRN-K577M; c) or exonuclease (WRN-E84A) activity (d), or treated with 0.2 μM aphidicolin. Arrows indicate chromosomal aberrations. (F) Frequency of fragile site FRA3B, FRA7H, and FRA16D expression in WS, WRN-E84A, WRN-K577M, and WSWRN cells. Samples were treated with different doses of aphidicolin and left until harvesting 24 h later. Frequency of fragile site induction is presented as the percentage of chromosome 3, 7, or 16 homologues with gaps and breaks at FRA3B, FRA7H, and FRA16D, respectively. Data are presented as means of three independent experiments. Asterisks indicate that the result is statistically significant compared with the wild type; P < 0.05 by t test. Error bars represent standard error. Bars, 2.5 μm.