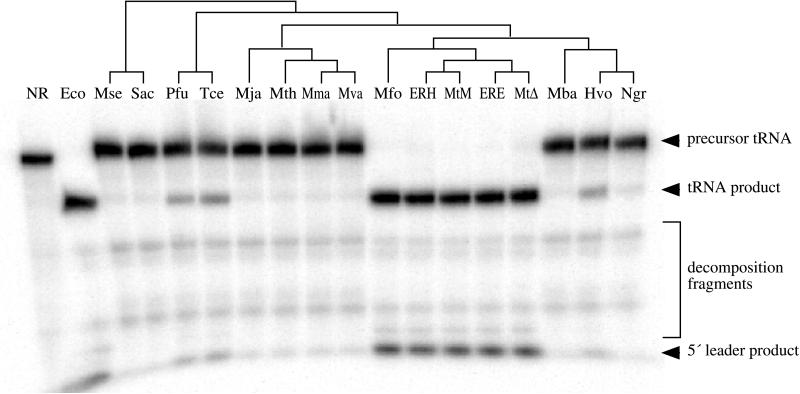
Figure 2.
Catalytic activity of synthetic archaeal RNase P RNAs. Assays contained 4 M ammonium acetate, 300 mM MgCl2, 50 mM Tris⋅Cl (pH 8), 0.1% SDS, 0.05% Nonidet P-40, 1.5 nM uniformly labeled B. subtilis pre-tRNAAsp, and ≈300 nM RNase P RNA (synthesized from cloned genes by transcription in vitro) and were incubated for 3.5 h at 45°C. The location of substrate and product bands are indicated. RNase P RNAs tested were no RNA (NR), E. coli (Eco), M. sedula (Mse), S. acidocaldarius (Sac), P. furiosus (Pfu), T. celer (Tce), M. jannaschii (Mja), M. thermolithotrophicus (Mth), Methanococcus marapaludis (Mma), Methanococcus vannielii (Mva), M. formicicum (Mfo), M. thermoautotrophicum ER-H (ERH), M. thermoautotrophicum Marburg (MtM), M. thermoautotrophicum ER-E (ERE), M. thermoautotrophicum ΔH (MtΔ), M. barkeri (Mba), H. volcanii (Hvo), and Natronobacterium gregoryi (Ngr). Activity by the N. gregoryi RNA is evident but weak. The phylogenetic relationships between these organisms based on small subunit ribosomal RNA sequences (26) are shown as a cladogram above the gel. Placement of the ER-E and ER-H RNAs, which were cloned from enrichment cultures inoculated with wastewater sludge, are based on similarity to the M. thermoautotrophicum strains ΔH and Marburg RNase P RNA sequences.