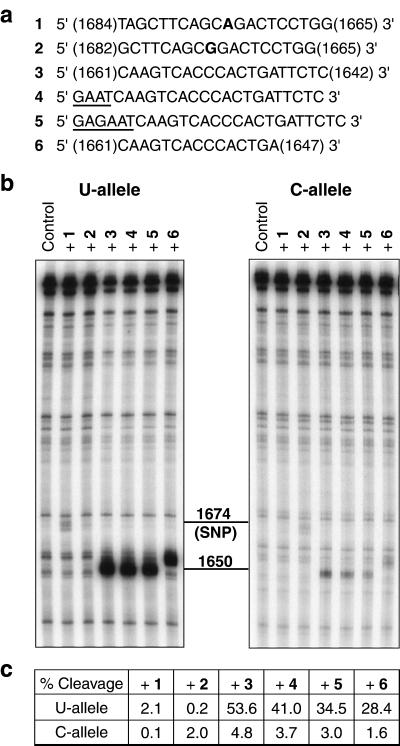
Figure 3.
E. coli RNase H digestion of both alleles of the 544-mer fragment of the RPA70–1,674 U/C mRNA. (a) Nucleotide sequence of the PS-oligos used in the assay. Oligos 1 and 2 target the polymorphic site; the bases opposing the polymorphism in the mRNA are shown in boldface. Oligos 3–6 target the site of different allelic structures around nucleotide 1,656. The terminal residues of oligos 1, 2, 3, and 6 are labeled according to the positions of the complementary nucleotides in RPA70 mRNA. Oligos 4 and 5 share the same target hybridization sequence as 3, but with additional nucleotides (shown underlined) added to facilitate folding into hairpin structures for intended enhancement in allele discrimination. (b) E. coli RNase H digestion patterns of the 5′-32P-labeled U and C alleles hybridized to the oligos in a. Cleavage sites were confirmed by running on the same gel RNase T1 digestion ladders for the two alleles (data not shown): they are centered at position 1,674 for oligos 1 and 2, 1,650 for oligos 3–5, and 1,654 for oligo 6. (c) Comparison of percent cleavages of the U and C alleles calculated from b. Oligo-dependent cleavage bands were integrated as a percentage of total RNA in each lane with the overlapping background-cleavage bands subtracted. Variations in sample loading to each lane were internally corrected by measuring percent cleavage instead of absolute band intensity.