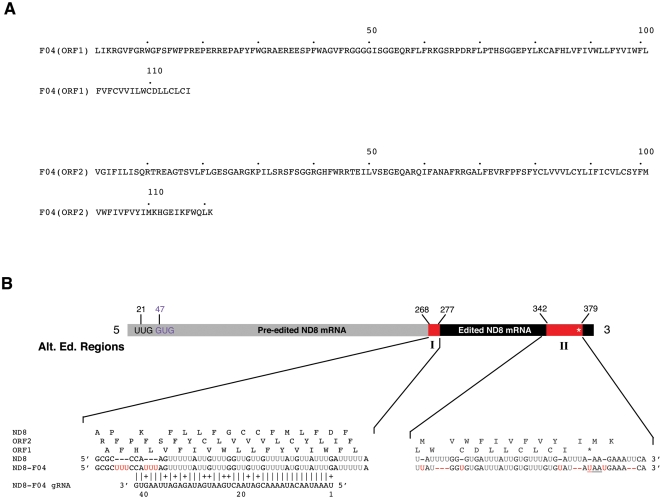
Figure 2. Alternative editing of ND8 mRNAs.
A) Protein sequence of ORF1 and ORF2, both sequences show no similarity to the bona fide ND8 (not shown). B) Bar depicts the mRNA sequence ND8-F04. Alignment of the RNA sequences for ND8, and ND8-F04 from the alternatively edited regions I, and II are shown below the bar. In black are the pre-edited residues, in grey the bona fide edited residues and in red are the alternatively edited residues from regions I–II. In black is the pre-edited, in grey the bona fide edited region and in red are the alternatively edited regions I and II. Alternative start codons UUG and GUG are shown in black and purple, respectively. Alignment of the RNA sequence of ND8 and cDNA ND8-F04 from the alternatively edited region I and II (red). Depicted above the RNA sequences are the corresponding amino acid sequences. Predicted gRNA for region I showing perfect complementarity (allowing for G:U) to ND8-F04 over 41 base pairs while having three mismatches to the ND8 sequence at positions 38 to 40 of the gRNA. Vertical bars indicate A:U or G:C base pairing; crosses indicates G:U base pairing. Red Us indicate alternatively inserted Us when compared with the bona fide ND8 sequence. Star depicts termination codon in the amino acid sequence. Underlined sequence depicts stop codon in the nucleotide sequence.