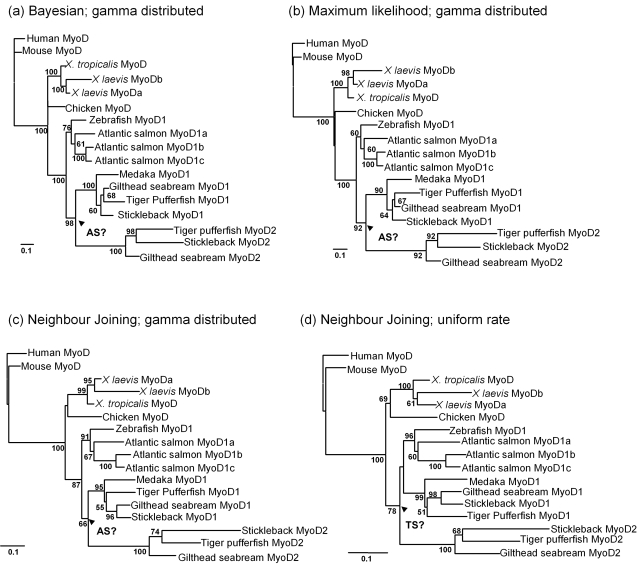
Figure 1. Unrooted phylograms of vertebrate MyoD amino acid sequences constructed using (a) Bayesian inference with a mixed model of amino acid substitutions and assuming a gamma distribution of among-site substitution rates (b) maximum likelihood with the WAG model of amino acid substitution and assuming a gamma distribution of among-site substitution rates (gamma distribution parameter estimated by PhyML to be 0.66) with 500 psuedobootstrap replicates for branch support (c) NJ with the Poisson correction model and assuming a gamma distribution of among-site rates (gamma distribution parameter = 0.66) and 1000 bootstrap replicates for branch support (d) NJ with the Poisson correction model assuming a uniform distribution of among-site substitutions rates with 1000 bootstrap replicates for branch support.
Arrows marked AS refer to the Acanthopterygian specific (AS) positioning of the teleost MyoD1/2 duplication inferred in trees a–c. The arrow marked TS shows the teleost specific (TS) positioning of the teleost MyoD1/2 duplication event in tree d. Scale bars show the number of substitutions per site. Branch confidence values >50% from the different reconstruction methods are shown.