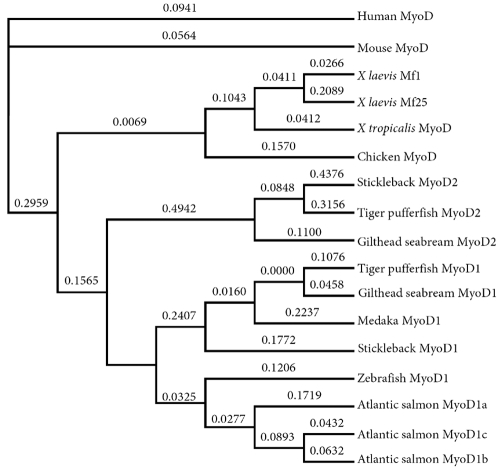
Figure 4. Unrooted ML cladogram of vertebrate MyoD amino acid sequences produced in PhyML [28] with an imposed ‘correct’ topology.
The amino acid alignment was the same as used in Fig. 1. The imposed ‘correct’ starting tree topology supported the teleost WGD event (Acanthopterygii MyoD2 branching internally to tetrapod MyoD sequences, but externally to teleost MyoD1 sequences) and PhyML was used to refine branch lengths only. The ‘correct’ topology for other MyoD duplication events (in X. Laevis and Atlantic salmon) was as observed in trees in Fig. 1a–d. Branch lengths (substitutions per site) are shown above each branch.