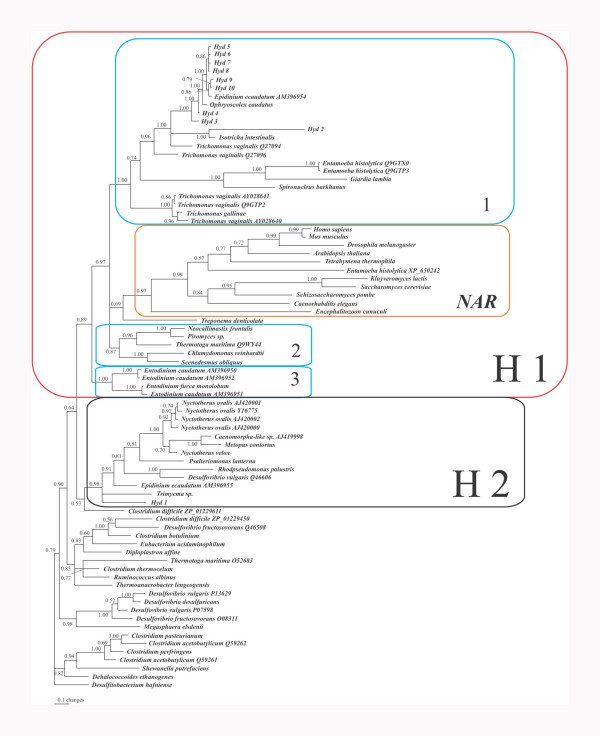
Figure 4.
Phylogenetic tree of the H-cluster of [FeFe]-hydrogenases and NARs or NARs-like proteins. Accession numbers of sequences are indicated when more than one sequence from a species is included. The numbers at the nodes represent the posterior probability resulting from a Bayesian inference. Hyd 1–10: H-clusters recovered from a metagenomic approach using DNA from total ciliate population in the rumen of a cow. The H1 block marks the "classical " [FeFe] hydrogenases and NAR's. Block 1 is characterized by the clade of Trichomonas vaginalis (long and short – type) hydrogenases. It hosts also the majority of the rumen sequences plus the hydrogenases from the type-strain rumen ciliates Epidinium ecaudatum, Ophryoscolex caudatus, and Isotricha intestinalis. Block 2 marks the long-type hydrogenases from the anaerobic chytridiomycetes Neocallimastix and Piromyces and the (short) plastidic hydrogenases from the algae Chlamydomonas and Scenedesmus. Block 3 marks H-clusters from rumen ciliates that are likely to lack hydrogenosomes. Block H2 marks a well supported clade of Fe hydrogenases dominated by N. ovalis. Besides N. ovalis and its close relatives, this clade consists of hydrogenases from the amoeboflagellate Psalteriomonas lanterna, the rumen ciliate Epidinium ecaudatum, the free-living ciliate Trimyema sp. and the rumen (meta) sequences Hyd 1. A fusion of the H-cluster with the 24 and 51 kDa modules has so far only been observed for the N. ovalis clade. The Psalteriomonas hydrogenase has no fused 24/51 kDa modules.