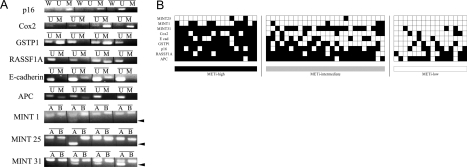
Figure 1.
Aberrant CpG island methylation profile of HCC. A: Representative results of methylation-specific PCR and combined bisulfite restriction enzyme analysis (COBRA). Bisulfite-modified DNA was amplified with primers specific to the methylated (M) or unmethylated (U) CpG islands of the p16, Cox2, GSTP1, RASSF1A, E-cadherin, and APC gene promoters. For the p16 gene promoter, we also performed PCR with primers (W) located outside the CpG islands to evaluate the efficiencies of the bisulfite modification; this PCR reaction never occurs if the modification is performed adequately. For the MINT 1, 25, and 31 clones, COBRA was performed in which only methylated samples were digested with restriction enzymes, and their locations are indicated by arrowheads. B: Before enzymatic digestion. A: After enzymatic digestion. B: Overview of the methylation analysis of nine CpG islands in 60 HCCs. Filled boxes indicate the presence of methylation and open boxes indicate the absence of methylation. The rows and columns indicate the nine CpG islands and the 60 HCC tumors, respectively.