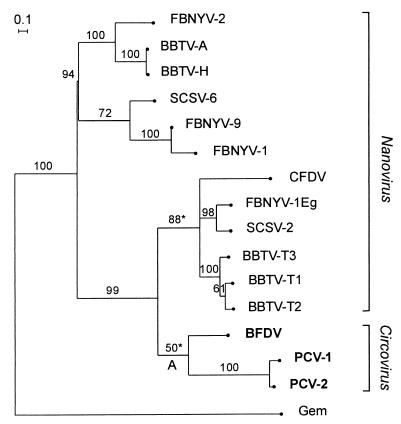
Figure 3.
A maximum likelihood tree for the N-terminal region of the available nanovirus and circovirus Rep amino acid sequences (up to position 129, see Fig. 1). The equivalent sequences from seven geminiviruses were used as an out-group (marked “Gem”) to root the tree. Sequences: BBTV-A and -H, Banana bunchy top nanovirus isolates from Australia and Hawaii; BBTV-T1, -T2, and -T3, Taiwanese isolates DNAs 1, 2, and 3; BFDV, Psittacine beak and feather disease circovirus; CFDV, Coconut foliar decay nanovirus; FBNYV-1, -2, -9, and -1Eg, Faba bean necrotic yellows nanovirus components from isolates from Syria and Egypt; PCV-1 and -2, Porcine circovirus types 1 and 2; SCSV-2 and -6, Subterranean clover stunt nanovirus components 2 and 6. Bootstrap values are percentages from 10,000 neighbor-joining trees inferred from the amino acid sequences. Asterisks mark branches not found in the maximum likelihood or most parsimonious trees inferred from nucleotide sequences. Note that several nanovirus isolates have two or more distinct Rep genes carried by different genomic molecules.