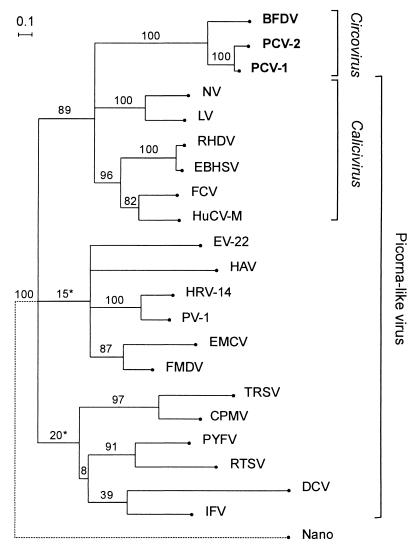
Figure 4.
A maximum likelihood tree for the 2C-protein amino acid conserved sequences of picorna-like viruses and the equivalent region from the circovirus Reps (from position 178 to 313; see Fig. 1). The equivalent C-terminal regions of six nanovirus Reps were included in the analysis and these sequences are represented by the node marked “Nano.” The actual estimate for the length of the branch leading to the “Nano” node is double that shown. Sequences: BFDV, Psittacine beak and feather disease circovirus; CPMV, Cowpea mosaic comovirus, DCV, Drosophila virus C; EBHSV, European brown hare syndrome calicivirus; EMV, Encephalomyocarditis cardiovirus; EV-22, Echovirus 22; FCV, Feline calicivirus; FMDV, Foot and mouth disease aphthovirus; HAV, Hepatitis A hepatovirus; HRV-14, Human rhinovirus 14; HuCV-M, Human calicivirus Manchester isolate; IFV, Infectious flacherie virus; LV, Lordsdale calicivirus; NV, Norwalk calicivirus; PCV-1 and -2, Porcine circovirus types 1 and 2; PV-1, Poliovirus 1; PYFV, Parsnip yellow fleck sequivirus; RHDV, Rabbit hemorrhagic disease virus; RTSV, Rice tungro spherical waikavirus; TRSV, Tobacco ringspot nepovirus. Bootstrap values are percentages from 10,000 neighbor-joining trees inferred from the amino acid sequences. Asterisks mark branches not found in the maximum likelihood or most parsimonious trees inferred from nucleotide sequences.