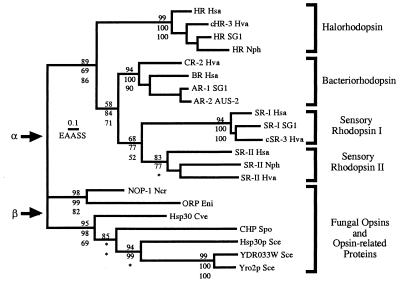
Figure 3.
Phylogenetic relationship between NOP-1 and related proteins from fungi and archaea. Phylogeny of the opsins was estimated by ML quartet puzzling, NJ of γ distances with α = 2, and MP. Analyses were conducted by using the complete alignment. Estimates of branch lengths were obtained by ML, assuming the PAM model of evolution with empirical amino acid frequencies, and the scale bar indicates 0.1 estimated amino acid substitutions per site (EAASS) by using this model. Support for the monophyly of subgroups of archaeal rhodopsins and all groupings within the fungal proteins is shown, with the quartet puzzling values above branches and the NJ and MP bootstrap proportions below branches. Cases in which alternative groupings were supported by NJ or parsimony are indicated by ∗ (within the fungal proteins, NJ and parsimony bootstrap consensus trees reverse the branching order of C. versicolor Hsp30 and the S. pombe conserved hypothetical protein). Two possible tree roots, α and β, are indicated by arrows. Cve, Coriolus versicolor; Eni, Emericella nidulans (the sexual form of Aspergillus nidulans); Hsa, Halobacterium salinarum; Hva, Haloarcula vallismortis; Ncr, Neurospora crassa; Nph, Natronomonas pharaonis; Sce, Saccharomyces cerevisiae; Spo, Schizosaccharomyces pombe. International Collaboration (IC) accession numbers: HR Hsa, P16102; cHR-3 Hva, P94853; HR SG1, P25964; HR Nph, P15647; CR-2 Hva, Q53496; BR Hsa, P02945; SR-1 SG1, P19585; AR-2 AUS-2, P29563; SR-I Hsa, P25964; SR-I SG1, P33743; cSR-3 Hva, Q48334; SR-II Hsa, P71411; SR-II Nph, P42196; SR-II Hva, P42197; NOP-1 Ncr, AF135863; ORP Eni, AA787158, AA785169, AA786492; Hsp30 Cve, AB003518; CHP Spo, AL031824; Hsp30p Sce, S31838; YDR033W Sce, S61586; Yro2p Sce, P38079.