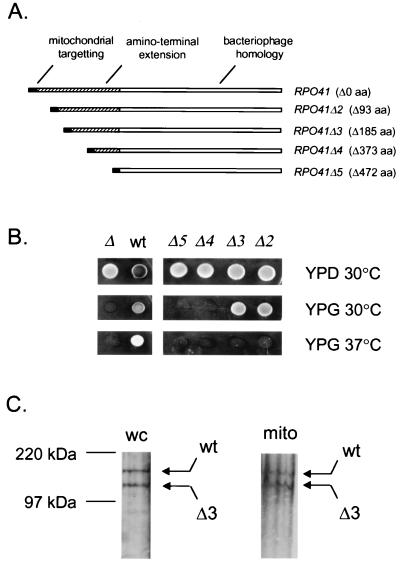
Figure 1.
Characterization of amino-terminal deletions in mtRNA polymerase. (A) A schematic representation of yeast mtRNA polymerase and deletion mutations used in this study is shown with important features labeled as follows: mitochondrial targeting sequence, black box; region containing bacteriophage homology, open box; amino-terminal extension, hatched box. The number of amino acids deleted (Δ) in each protein is given in parentheses. (B) Phenotypic analysis of RPO41 deletion mutants by plasmid shuffle. Mitochondrial function was assessed in each strain by comparing growth on YPD and YPG at 30°C and 37°C. The RPO41 genotype of each strain after plasmid shuffle is given as follows: Δ, rpo41; wt, RPO41; Δ2-Δ5, rpo41Δ2-Δ5. (C) Western analysis of rpo41Δ3-encoded mtRNA polymerase from GS129. After growth at 37°C for ≈20 generations, protein from whole cells (wc) and from a purified mitochondrial fraction (mito) were analyzed. Signals for the wild-type (wt) and rpo41Δ3-encoded (Δ3) protein are indicated with arrows. Protein molecular mass standards are indicated on the left.