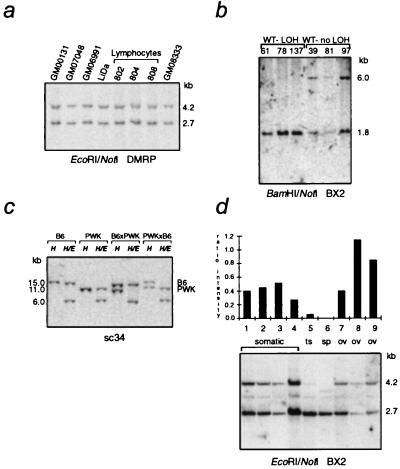
Figure 3.
Differential methylation of KvDMR1. (a) DNA isolated from normal lymphoblastoid (GM00131, GM07048, and GM06991), peripheral blood lymphocyte (802, 804, and 808), or fibroblast (GM08333) cells was digested with EcoRI and NotI and hybridized with DMRP (see Fig. 2a). (b) Southern blot of BamHI/NotI-digested DNA from Wilms’ tumors (WT) with loss of heterozygosity (LOH) or without LOH (no LOH) in 11p15 and hybridized with the BX2 probe (Fig. 2a). (c) Hybridization of sc34 to HindIII (H) and HindIII/EagI (H/E) double digests of adult kidney DNA from reciprocal [C57BL/6J (B6) × PWK]F1 animals (for F1 hybrids, the maternal parent is specified first). (d) The BX2 probe was hybridized to EcoRI/NotI digests of human DNA from somatic tissues (peripheral blood lymphocytes or brain), testes (ts), sperm (sp), and fetal ovaries (ov). The ratio of the intensity of the upper and lower bands is shown in the accompanying histogram. The faint band at 3–3.5 kb present in all lanes is a cross-hybridizing locus observed when experiments are done at reduced stringency. The additional band seen in the sperm lane is likely due to a heterogeneous methylation at this cross-reacting locus.