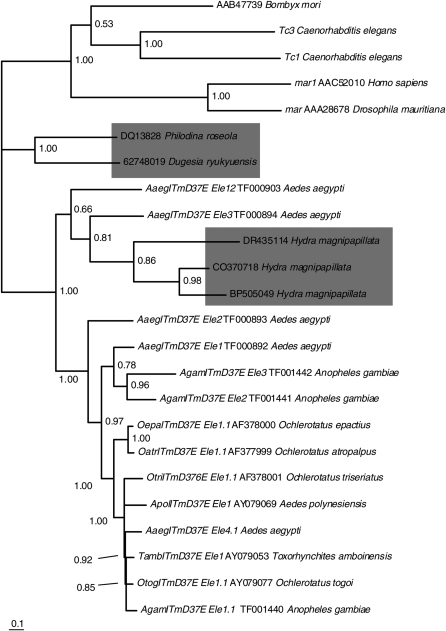
Figure 1.—
ITmD37E TEs are of an ancient origin. Consensus tree (>50%) based on conceptual translations constructed using MrBayes version 3.1.2 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003). ITmD37E sequences from freshwater invertebrates are in shaded boxes. Tc1, mariner, and DD37D (Bmmar1) TEs are used to root the tree. Clade credibility values are shown at each node. The scale represents substitutions per site. All mosquito elements have intact ORFs except for AaegITmD37E_Ele4.1, AgamITmD37E_Ele2, and AgamITmD37E_Ele3, which were obtained from whole-genome sequence projects. Element name, accession number, and species name are given when applicable. See supplemental File S1 (http://www.genetics.org/supplemental/) for methods. Information pertaining to sequences can be found in supplemental File S2. See supplemental File S3 for alignment.