Joint X-ray and neutron crystallographic data have been collected from the oligonucleotide d(CGCGCG) crystallized without polyamine and at low pH in order to study hydration in the protein-binding major groove of Z-DNA.
Keywords: d(CGCGCG), Z-DNA, hydration
Abstract
In order to crystallographically study the hydration of the major groove (convex surface) of Z-DNA, the oligonucleotide d(CGCGCG) has been synthesized. Single crystals were grown by vapor diffusion using the hanging-drop and sitting-drop methods for X-ray studies and by batch crystallization and evaporation within silicon tubes for neutron studies. Hexagonal crystals were obtained without the use of duplex-stabilizing polyamines and at an acid pH. X-ray data collected at room temperature (1.5 Å resolution; unit-cell parameters a = 17.90, b = 30.59, c = 44.61 Å) and at 100 K (1 Å resolution; a = 17.99, b = 30.98, c = 44.07 Å) and neutron data collected at room temperature (1.6 Å resolution; a = 18.00, b = 31.16, c = 44.88 Å) indicate that the DNA is in the Z-form packing in space group P212121.
1. Introduction
X-ray fiber diffraction has shown that hydration plays a central role in stabilizing the different forms of the DNA duplex (A, B, C, D and Z) and facilitating transitions between them (Leslie et al., 1980 ▶). Although subsequent X-ray studies of oligonucleotide single crystals have provided a wealth of information on DNA hydration (Dickerson et al., 1982 ▶, Kennard & Hunter, 1991 ▶; Berman, 1994 ▶), they have been limited by the absence of H atoms from electron-density maps. Water molecules appear as single peaks corresponding to the positions of O atoms and are difficult to differentiate from peaks arising from other solvent atoms even at the subatomic resolution of 0.6 Å (Tereshko et al., 2001 ▶). Neutron crystallography is ideal for studying hydration because H atoms and in particular their isotope D scatter neutrons efficiently. The identification of all three atoms in an ordered water molecule using neutron crystallography and H2O/D2O substitution leads to the determination of its hydrogen-bonding coordination. The combination of information from X-ray and neutron crystallography can be more powerful than either technique on its own because of the potential to investigate the nature of disorder and to differentiate water from other solvent molecules.
The available neutron beams are weak compared with X-ray beams and therefore larger sample sizes are required. Although it is often difficult to grow large single crystals of oligonucleotide, it is straightforward to assemble large bundles of DNA fibers, an advantage that has allowed the distributions of localized water around the A, B and D forms of DNA to be determined by neutron fiber diffraction (Langan et al., 1992 ▶; Shotton et al., 1997 ▶; Forsyth et al., 1989 ▶). These studies have provided some unique insights into the role of water in stabilizing DNA (Fuller et al., 2004 ▶; Langan, 2005 ▶), but have been limited in their resolution (∼3 Å).
Recent advances in instrumentation and crystallization techniques now make possible neutron crystallography studies of oligonucleotides (Arai et al., 2002 ▶, 2005 ▶; Chatake et al., 2005 ▶). In order to reveal the detailed hydration of Z-DNA, Chatake et al. (2005 ▶) collected data from large (1.6 mm3) crystals of d(CGCGCG) cocrystallized with spermine and MgCl2. The Z form, unlike the other forms of DNA, is left-handed. Although the biological function of Z-DNA is not completely clear, it has been associated with a number of binding proteins that have specific and water-mediated interactions with the major groove of Z-DNA (Ha et al., 2004 ▶).
An important result from the study of Chatake et al. (2005 ▶) is that water molecules form an ordered network of hydrogen bonds in the minor groove that helps stabilize Z-DNA. However, a spermine cation was found in the major groove, bound to the DNA, and water in that groove was rotationally disordered. The aim of the work presented here is to obtain crystals of Z-DNA with no spermine present in the major groove and to subject them to joint X-ray and neutron crystallographic analysis. In this way, we hope to gain insights into how water is reorganized or displaced on protein binding and what supporting role, if any, major-groove hydration plays to minor-groove hydration in stabilizing Z-DNA. We wished to collect X-ray and neutron data for joint analysis at room temperature in order to provide the most physiologically relevant results. However, we also wished to collect a high-resolution X-ray data set at cryo-temperature in order to assess the different levels of information available from a high-resolution X-ray crystallographic structure compared with a medium-resolution joint X-ray and neutron crystallographic structure.
2. Materials and methods
2.1. Synthesis and purification
An Applied Biosystems ABI four-channel oligonucleotide synthesizer using the solid-phase (CPG) phosphoramidite approach was used to synthesize d(CGCGCG). The reaction was about 90% efficient per base addition. After synthesis, oligonucleotides were cleaved from the support, deprotected and then detritylated using standard methods. A few drops of ammonia were added to the final sample after elution from the column (Cruachem OPC cartridges) with 20% acetonitrile in HPLC-grade H2O and the sample was then lyophilized. At a later date, the sample was suspended in HPLC-grade H2O or D2O to a concentration of about 20 mg ml−1 and then, in order to anneal the duplex, heated to 353 K for 1 h and left to cool to room temperature overnight in a styrofoam box. The sample was stored at 273 K without further purification or the addition of duplex-stabilizing polyamines or divalent cations.
2.2. Crystallization
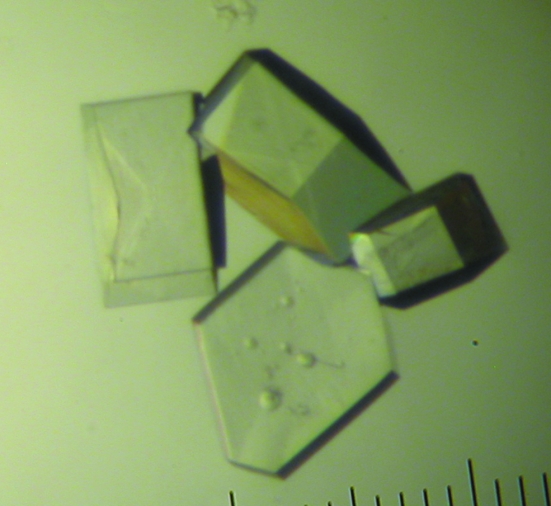
Initial crystallization conditions were identified by the hanging-drop diffusion method at 293 K using the Natrix screen (Hampton Research), crystallization plates and siliconized cover slides; drops contained 1 µl reservoir solution and 1 µl sample and were equilibrated against 500 µl reservoir solution. Crystals appeared within days over a wide variety of conditions. The screen was repeated a number of times and the largest crystals (reaching dimensions of over 300 µm in length in about 2 d and hexagonal in shape) were consistently obtained with Natrix solution No. 2 (0.01 M magnesium acetate, 0.05 M MES pH 5.6, 2.5 M ammonium sulfate) (Fig. 1 ▶).
Figure 1.
Crystals of d(CGCGCG) used for X-ray crystallography, with a maximum dimension of ∼300 µm, obtained from a 2 µl hanging drop with 0.01 M magnesium acetate, 0.05 M MES pH 5.6, 2.5 M ammonium reservoir.
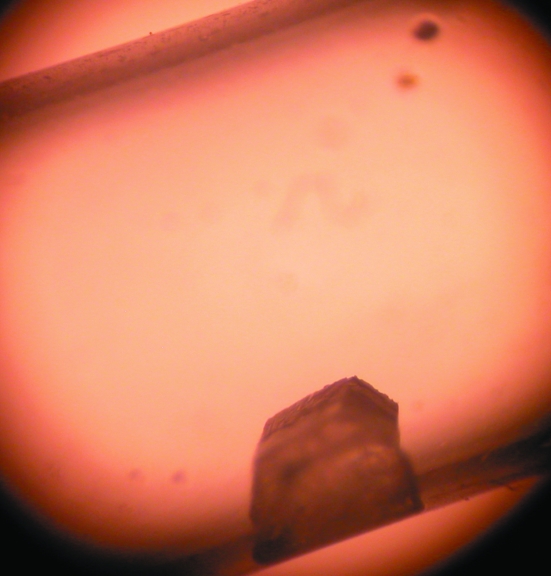
Crystals over 1 mm in length were obtained using both the hanging-drop and sitting-drop vapor-diffusion methods and Natrix solution No. 2 with sample concentrations of up to 50 mg ml−1 and drops of up to 20 µl in size at 293 K. However, attempts to mount these in capillary tubes failed because the crystals fractured when moved. Batch crystallization within synthetic fused silica tubes (VitroCom Inc.; internal diameter 3 mm), which are less likely to break during handling than thin-walled quartz capillary tubes and are relatively transparent to neutrons, was therefore attempted using a range of different proportions of sample and Natrix solution 2. One silica tube containing 100 µl solution No. 2 and 100 µl sample (50 mg ml−1) and plugged at both ends with beeswax, each plug having a small hole in it (200 µm in diameter) to allow slow evaporation, was left in a styrofoam box. Over several weeks, a large colorless transparent hexagonal crystal (∼0.7 mm3) was formed, leaving only a small amount of batch solution (∼20 µl) at one end of the silicon tube (Fig. 2 ▶).
Figure 2.
A crystal of d(CGCGCG) used for neutron crystallography with a maximum dimension of ∼900 µm obtained by batch crystallization within a fused silica tube (3 mm outer diameter).
2.3. Deuteration
Isotopic H/D exchange (deuteration) involves replacing H atoms in the crystal by D atoms and is commonly used in neutron crystallography to reduce background scattering and to increase the effective neutron scattering power of crystals by up to an order of magnitude. Isotopic exchange of labile accessible H atoms can be achieved by vapor diffusion. A deuterated version of solution No. 2 was prepared using D2O rather than H2O and perdeuterated ammonium sulfate rather than its hydrogenous equivalent. Because of the relatively small amount of MES (2-morpholinoethanesulfonic acid monohydrate; C6H13NO4S) in solution No. 2 we did not use a perdeuterated equivalent. The crystal for neutron studies was deuterated by replacing the mother liquor in the silicon tube by a small amount (20 µl) of the deuterated solution No. 2, both ends of the tube were sealed and the tube was left overnight. This H2O/D2O vapor-diffusion exchange process was repeated five times over 3 d. The appearance of the crystal did not change. Deuterated crystals for X-ray studies were prepared by growing fresh crystals in hanging drops using deuterated solutions and reagents.
2.4. Data collection
X-ray diffraction data were measured at low and room temperature using a Riguku FR-E rotating-anode generator with Osmic Blue focusing optics and a Saturn 92 CCD detector. Crystals were mounted on cryoloops; however, for room-temperature data collection the crystal-mount assembly was placed inside a microRT plastic sleeve (MiTeGen) with a droplet of mother liquor at the top to stabilize the crystal during diffraction studies. The crystal for low-temperature studies was flash-cooled in cold helium at 40 K with a CryoIndustries Inc. CF-4 cryostat and then removed to the X-ray beam and cooled during data measurement with an Oxford Cryosystems 700 nitrogen cryostat. The temperature at the crystal position was 112 K.
For both room- and low-temperature data measurement, the detector was placed at a 2θ angle of 45°, with 10 s exposures of 0.5° oscillation width per frame. Four 120° ω-scan passes were made with changes made in either the χ- or ϕ-axis orientation to complete reciprocal-space sampling. Radiation damage limited the number of usable frames and completeness of the room-temperature data. Data were integrated using d*TREK (Pflugrath, 1999 ▶) from the CrystalClear 1.3.6 software package (Rigaku).
Neutron diffraction data were collected at room temperature on the PCS at LANSCE (Langan et al., 2004 ▶), integrated using a version of d*TREK (Pflugrath, 1999 ▶) modified for wavelength-resolved neutron Laue protein crystallography (Langan & Greene, 2004 ▶), wavelength-normalized using LAUENORM (Helliwell et al., 1989 ▶) and then merged using SCALA (Evans, 1997 ▶) using previously described strategies (Hanson et al., 2004 ▶; Li et al., 2004 ▶; Sukumar et al., 2005 ▶) (Table 1 ▶).
Table 1. Experimental details.
| Neutron | X-ray | ||
|---|---|---|---|
| Ambient | Ambient | 100 K | |
| Crystal data | |||
| Space group | P212121 | P212121 | P212121 |
| Unit-cell parameters (Å) | |||
| a | 18.00 (6) | 17.90 (5) | 17.99 (5) |
| b | 31.16 (12) | 30.59 (5) | 30.98 (5) |
| c | 44.88 (13) | 44.61 (5) | 44.07 (5) |
| Crystal volume (mm3) | 0.7 | 0.07 | 0.1 |
| Data collection | |||
| Nominal resolution (Å) | 1.6 (1.69–1.60) | 1.5 (1.55–1.50) | 1.1 (1.13–1.09) |
| Unique reflections | 2171 | 3549 | 10138 |
| Multiplicity | 2.6 (2.0) | 2.4 (2.5) | 5.5 (3.7) |
| Completeness (%) | 62 (51) | 84 (76) | 93.7 (85.9) |
| Rmerge (%) | 4.9 (27.5) | 7.9 (31.7) | 7.4 (20.8) |
| Diffractometer | PCS | FR-E | FR-E |
| λ (Å) | 0.6–6 | Cu Kα | Cu Kα |
| I/σ(I) | 5.2 (2.5) | 18.9 (4.3) | 34.2 (8.8) |
3. Results and discussion
The 11 crystal structures of d(CGCGCG) in the Nucleic Acid Database (NDB) have the same P212121 space group but fall into two families that differ in the absolute orientation of the duplexes in the unit cell and therefore the exact unit-cell parameters. One family (A) consists of ZDF053, ZDF052, ZDF001, ZDF002, DDF027, ZD0003 and ZD0005 and has average unit-cell parameters a = 17.93 (5), b = 31.35 (20), c = 44.59 (12) Å. The other family (B) consists of ZDF035, ZDF029, ZD0012 (the neutron structure of Chatake et al., 2005 ▶) and ZD0004 and has average unit-cell parameters a = 18.36 (9), b = 30.72 (5), c = 42.82 (40) Å. The values of the unit-cell parameters reported here indicate that our structure belongs to family A and this has been confirmed by molecular replacement. It should be emphasized that only one of the 11 NDB entries is free from polyamines and the data reported here are the first from a joint X-ray and neutron study of a polyamine-free form. At ambient temperature the lattice constants of the crystal for neutron data collection are all slightly larger than those of the crystal for X-ray data collection, by as much as four experimental standard deviations. The ambient temperatures at Los Alamos during the neutron experiment and at Toledo during the X-ray experiment were most likely to have been quite different.
A number of methods have been previously reported for growing crystals inside capillaries, including vapor diffusion (McPherson, 1999 ▶), microdialysis (Zeppezauer et al., 1968 ▶), liquid–liquid or free-interface diffusion (Salemme, 1972 ▶) and gel acupuncture (García-Ruiz & Moreno, 1994 ▶). We believe that this is the first report of crystallization within a capillary using evaporation as a means of obtaining large crystals for neutron crystallography, although a method has been reported for crystal growth in X-ray-transparent permeable plastic tubes for high-throughput X-ray crystallography (Kalinin & Thorne, 2005 ▶). We are currently investigating ways of refining our crystallization procedure in order to provide a general method for growing large crystals to ease handling for neutron crystallography.
Acknowledgments
The PCS is funded by the Office of Science and the Office of Biological and Environmental Research of the US Department of Energy. PL and MM thank NIH–NIGMS (1R01GM071939-01) for support. BLH thanks NSF MCB-0446218 for support.
References
- Arai, S., Chatake, T., Minezaki, Y. & Niimura, N. (2002). Acta Cryst. D58, 151–153. [DOI] [PubMed] [Google Scholar]
- Arai, S., Chatake, T., Ohhara, T., Kurihara, K., Tanaka, I., Suzuki, N., Fujimoto, Z., Mizuno, H. & Niimura, N. (2005). Nucleic Acids Res.33, 3017–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, H. M. (1994). Curr. Opin. Struct. Biol.4, 345–350.
- Chatake, T., Tanaka, I., Umino, H., Aria, S. & Niimura, N. (2005). Acta Cryst. D61, 1088–1098. [DOI] [PubMed] [Google Scholar]
- Dickerson, R. E., Drew, H. R., Conner, B. N., Kopka, M. L. & Pjura, P. E. (1982). Cold Spring Harbor Symp. Quant. Biol.47, 13–24. [DOI] [PubMed]
- Evans, P. (1997). Proceedings of the CCP4 Study Weekend. Recent Advances in Phasing, edited by K. S. Wilson, G. Davies, A. W. Ashton & S. Bailey, pp. 97–102. Warrington: Daresbury Laboratory,
- Forsyth, V. T., Mahendrasingam, A., Langan, P., Pigram, W. J., Stevens, E. D., Al-Hayalee, Y., Bellamy, K. A., Greenall, R. J., Mason, S. A. & Fuller, W. (1989). Inst. Phys. Conf. Ser.101, 237–242.
- Fuller, W., Forsyth, V. T. & Mahendrasingam, A. (2004). Philos. Trans. R. Soc. Lond. B Biol. Sci.359, 1237–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Ruiz, J. M. & Moreno, A. (1994). Acta Cryst. D50, 484–490. [DOI] [PubMed] [Google Scholar]
- Ha, S. C., Lokanath, N. K., Quyen, D. V., Wu, C. A., Lowenhaupt, K., Rich, A., Kim, Y.-G. & Kim, K. K. (2004). Proc. Natl Acad. Sci. USA, 101, 2712–2717. [Google Scholar]
- Hanson, B. L., Langan, P., Katz, A. K., Li, X., Harp, J. M., Glusker, J. P., Schoenborn, B. P. & Bunick, G. J. (2004). Acta Cryst. D60, 241–249. [DOI] [PubMed] [Google Scholar]
- Helliwell, J. R., Habash, J., Cruickshank, D. W. J., Harding, M. M., Greenhough, T. J., Campbell, J. W., Clifton, I. J., Elder, M., Machin, P. A., Papiz, M. Z. & Zurek, S. (1989). J. Appl. Cryst.22, 483–497. [Google Scholar]
- Kalinin, Y. & Thorne, R. (2005). Acta Cryst. D61, 1528–1532. [DOI] [PubMed] [Google Scholar]
- Kennard, O. & Hunter, W. N. (1991). Angew. Chem. Int.30, 1254–1277.
- Langan, P. (2005). Crystallogr. Rev.11, 125–147.
- Langan, P., Forsyth, V. T., Mahendrasingam, A., Pigram, W. J., Mason, S. A. & Fuller, W. (1992). J. Biomol. Struct. Dyn.10, 489–503. [DOI] [PubMed] [Google Scholar]
- Langan, P. & Greene, G. (2004). J. Appl. Cryst.37, 253–257. [Google Scholar]
- Langan, P., Greene, G. & Schoenborn, B. P. (2004). J. Appl. Cryst.37, 24–31. [Google Scholar]
- Leslie, A. G. W., Arnott, S., Chandrasekaran, R. & Ratliff, R. L. (1980). J. Mol. Biol.143, 49–72. [DOI] [PubMed] [Google Scholar]
- Li, X., Langan, P., Bau, R., Tsyba, F., Jenney, F. E. Jr, Adams, M. W. W. & Schoenborn, B. P. (2004). Acta Cryst. D60, 200–202. [DOI] [PubMed] [Google Scholar]
- McPherson, A. (1999) Crystallization of Biological Macromolecules, pp. 193–194. Cold Spring Harbor: Cold Spring Harbor Laboratory Press.
- Pflugrath, J. W. (1999). Acta Cryst. D55, 1718–1725. [DOI] [PubMed] [Google Scholar]
- Salemme, F. R. (1972). Arch. Biochem. Biophys.151, 533–539. [DOI] [PubMed] [Google Scholar]
- Shotton, M. W., Pope, L. H., Forsyth, V. T., Langan, P., Grimm, H., Rupprecht, A., Denny, R. C. & Fuller, W. (1997). Physica B, 241–243, 1156–1158.
- Sukumar, N., Langan, P., Mathews, F. S., Jones, L. H., Thiyagarajan, P., Schoenborn, B. P. & Davidson, V. L. (2005). Acta Cryst. D61, 640–642. [DOI] [PubMed] [Google Scholar]
- Tereshko, V., Wilds, C. J., Minasov, G., Prakash, T. P., Maier, M. A., Howard, A., Wawrzak, Z., Manoharan, M. & Egli, M. (2001). Nucleic Acids Res.9, 5443–5457. [DOI] [PMC free article] [PubMed]
- Zeppezauer, M., Eklund, H. & Zeppezauer, E. S. (1968). Arch. Biochem. Biophys.126, 564–573. [DOI] [PubMed] [Google Scholar]