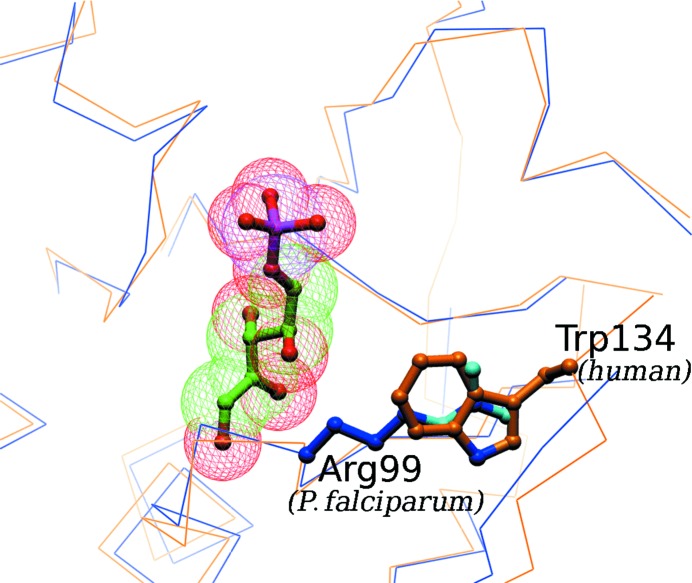
Figure 3.
Potentially exploitable difference in the ribose-5-phosphate-binding site in the P. falciparum and human RpiA proteins. The blue backbone trace in the figure is that of the P. falciparum structure presented here; the gold trace is that of a homology model for the human RpiA based on the crystal structure (1xtz) of the yeast homolog. The ribose-5-phosphate substrate is positioned as seen in the T. thermophilus enzyme–substrate complex (1uj5). The very different binding-site environment to the right of the substrate, Arg99 in P. falciparum and Trp134 in humans (Trp157 in yeast), suggests that it would be possible to design inhibitors with very different affinities for the two homologous enzymes.