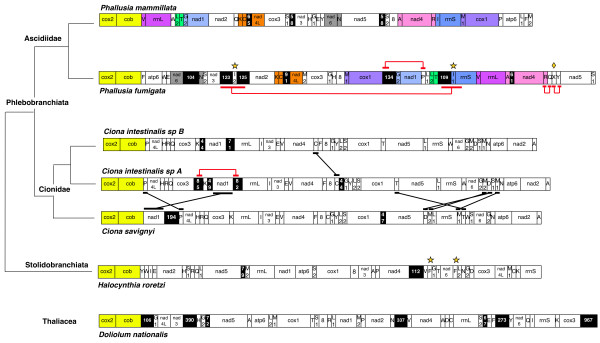
Figure 1.
Gene organization of tunicate mitochondrial genomes, and taxonomic classification of the analysed species. Gene blocks conserved between P. mammillata and P. fumigata are reported in different colours, with yellow indicating the gene pair conserved in all analysed tunicates. Sequences almost identical in the same mtDNA are underlined and linked by a red line. Genes transposed among the three Ciona species are underlined and linked by a black line. Non-coding (NC) regions equal or longer that 40 bp are indicated by black background, with numbers corresponding to their size (in bp). Abbreviations for protein-coding and rRNA genes are as in the main text, except for atp8 gene (abbreviation: 8). Transfer RNA genes are indicated according to the transported amino acid, except for: G1: Gly(AGR); G2: Gly(GGN); L1: Leu(UUR); L2: Leu(CUN); M1: Met(AUG); M2: Met(AUA); S1: Ser(AGY); S2: Ser(UCN). Diamond: trnX gene of P. fumigata. Star: intra-genome duplicated genes, that is trnI genes in P. fumigata (named I1 and I2), and trnF in Halocynthia roretzi. Feature table of Halocynthia roretzi mtDNA is as reported in [38].