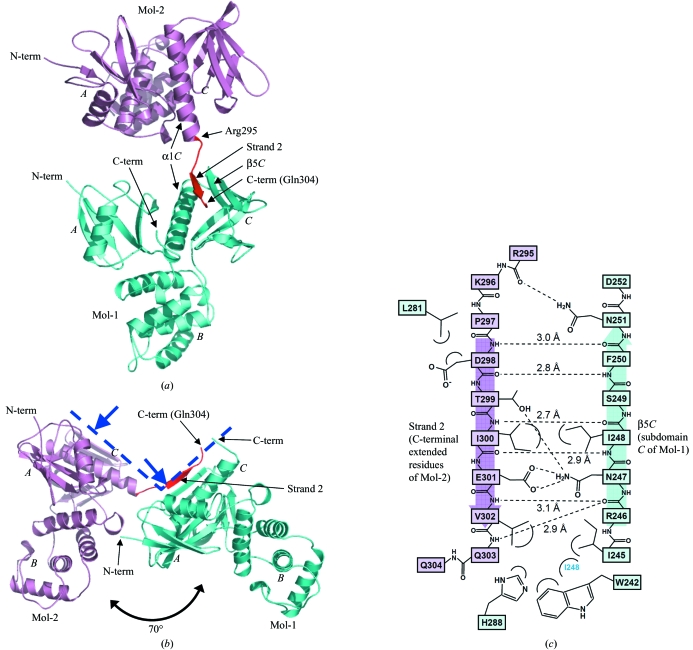
Figure 1.
Crystal structure of dimerized radixin FERM domains. (a) Overall structure as a ribbon model. Mol-1 is shown in light blue and Mol-2 in pink, with its C-terminal extended residues (Arg295–Gln304) in red. The radixin FERM domain consists of subdomains A (the N-terminal 82 residues), B (residues 96–119) and C (residues 204–297). (b) A side view shows that Mol-1 and Mol-2 are aligned in a similar orientation. The blue dashed line represents the membrane-interaction surface of a dimer and blue arrows indicate binding sites to phosphatidylinositol 4,5-bisphosphate (PIP2; Hamada, Shimizu et al., 2000 ▶). Figures were prepared using PyMOL (http://pymol.sourceforge.net). (c) Schematic representation of the interactions between the C-terminal extended residues (strand 2 in pink) and subdomain C (light blue). Hydrogen bonds are shown as dashed lines and distances are indicated.