Table 1. Crystallographic data for the dimerized radixin FERM domains.
Values in parentheses are for the outer resolution shell.
| X-ray data | |
| Space group | P21 |
| Unit-cell parameters (Å, °) | a = 57.2, b = 70.2, c = 110.8, β = 99.0 |
| Resolution (Å) | 30–2.8 (2.9–2.8) |
| Mosaicity (°) | 0.2–0.5 |
| Reflections (total/unique) | 68724/19703 |
| Completeness (%) | 96.4 (76.9) |
| 〈I/σ(I)〉 | 11.0 (1.5) |
| Rmerge (%) | 8.7 (56.4) |
| Refinement | |
| No. of residues included | 297 (Mol-1), 302 (Mol-2) (of 310) |
| No. of atoms | 5018 |
| Rwork/Rfree† (%) | 22.2/26.9 |
| Ramachandran plot (%) | |
| Most favoured | 83.7 |
| Additional allowed | 14.4 |
| Generously allowed | 1.3 |
| Disallowed | 0.6 |
| Average B factor (Å2) | |
| Mol-1 | 69.5 |
| Mol-2 (FERM/C-term 295–304) | 81.1/88.3 |
| R.m.s. bond length (Å) | 0.008 |
| R.m.s. bond angles (°) | 1.3 |
R
work = 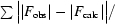
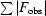 . R
free is the same as R
work except that it was calculated for a 5% subset of all reflections that were never used in crystallographic refinement.
. R
free is the same as R
work except that it was calculated for a 5% subset of all reflections that were never used in crystallographic refinement.
