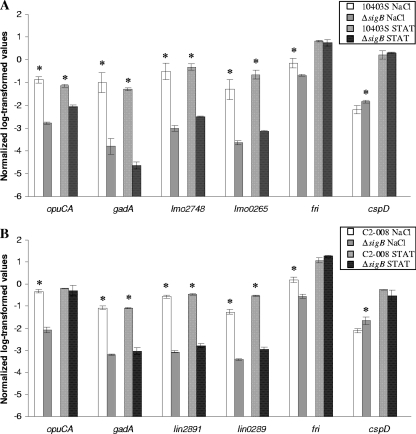
FIG. 1.
qRT-PCR confirmation of selected genes identified by microarray data as σB dependent in L. monocytogenes 10403S (A) and in L. innocua FSL C2-008 (B). qRT-PCRs were designed to be specific for the six selected L. monocytogenes and L. innocua genes (see Table S8 in the supplemental material); correlation plots for microarray and qRT-PCR results are provided as Fig. S8 in the supplemental material. The six genes were selected to represent genes with homologues in both species; homologous genes are presented in the same order in each figure, e.g., lmo2748 and lin2891 are homologous. Transcript levels for the test genes were normalized to the geometric mean of the transcript levels for housekeeping genes rpoB and gap; an asterisk indicates that normalized transcript levels for the parent and ΔsigB strain differed significantly for cells exposed to a given stress condition (salt stress or stationary phase). As microarray analyses indicated that transcript levels for L. innocua gap were lower in the ΔsigB strain, statistical analyses of transcript levels for L. innocua genes were also repeated using transcript levels normalized to rpoB only; except for cspD (see text), these statistical analyses identified the same differences as significant as the analyses based on transcript levels normalized to the geometric mean of rpoB and gap transcript levels.