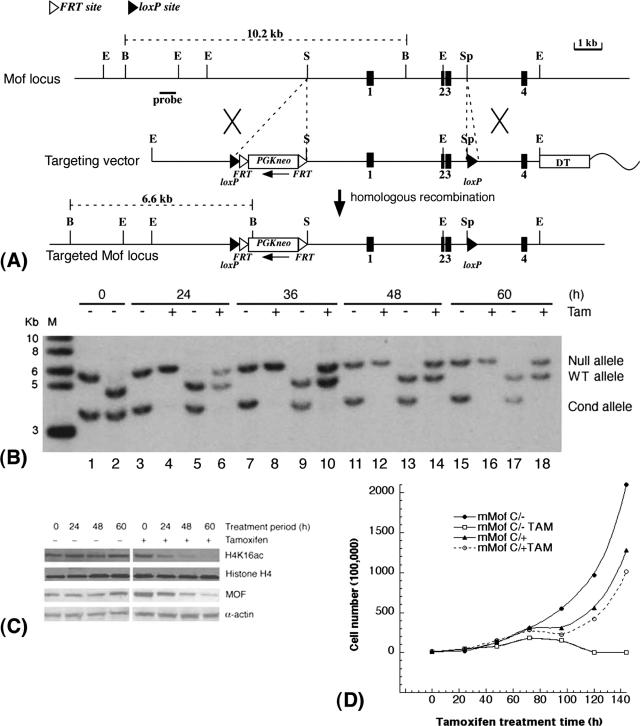
FIG. 5.
mMof is essential for proliferation and/or viability of ES cells. (A) Conditional inactivation of mMof. A partial restriction map of the genomic region containing mMof exons 1 to 4 (black boxes numbered 1 to 4) is shown at the top, followed by a diagram of the targeting vector used for insertion of the FRT-flanked neomycin cassette and loxP sites into the mMof gene (bottom). A diphtheria toxin A gene cassette is also included in the targeting construct as a negative selection marker. The wavy line represents plasmid sequence. The restriction sites shown are BglII (B), EcoRI (E), SalI (S), and SphI (Sp). (B) Southern blot analysis showing cre-mediated recombination of the floxed allele in mMofflox/+/Rosa26creERT2/+ and mMofΔflox/flox/Rosa26creERT2/+ ES cells after tamoxifen treatment. Genomic DNA was prepared from mMofΔflox/flox/Rosa26creERT2/+ (lanes 1, 3, 4, 7, 8, 11, 12, 15, and 16) or mMofflox/+/Rosa26creERT2/+ ES cells (lanes 2, 5, 6, 9, 10, 13, 14, 17, and 18) collected at 0, 24, 36, 48, and 60 h from cells treated with 200 nM tamoxifen (Tam) or not treated with tamoxifen (−), digested with PstI, and probed with the 5′ external probe. Note that after 24 h, the conditional (Cond) Mofflox allele is fully recombined in the presence of tamoxifen. The positions of molecular size markers in lane M (in kilobases) are indicated to the left of the gel. WT, wild type. (C) Western blot analysis for MOF and H4K16ac levels in mMofΔflox/flox/Rosa26creERT2/+ ES cells harvested at 0, 24, 48, and 60 h from cells treated with tamoxifen (Tam) (+) or not treated with tamoxifen (−). (D) Proliferation of mMofflox/+/Rosa26creERT2/+ and mMofΔflox/flox/Rosa26creERT2/+ ES cells with or without tamoxifen treatment. Cell numbers were determined at different time points to determine the population doublings in cells with and without mMof.