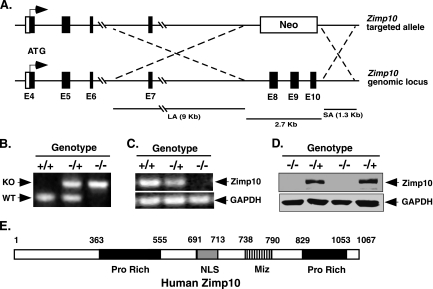
FIG. 2.
Targeted disruption of the Zimp10 gene. (A) Schematic representation of the Zimp10 targeting strategy. The Zimp10 locus was disrupted with a targeting vector that replaced approximately 2.7 kb of the Zimp10 genomic sequence containing exons 8 to 10 with a neomycin (Neo) resistance gene. The top line represents the neomycin targeting vector, and the bottom line represents the Zimp10 genomic sequence. The ATG translational start site is indicated with arrows. LA, long homologous recombination arm; SA, short homologous recombination arm. (B) PCR-based genotyping of E9.5 embryos using primers specific for the wild-type or neomycin-targeted allele. Knockout (KO) and wild-type (WT) bands are indicated. (C) RT-PCR analysis of Zimp10 transcripts in E9.5 embryos using primers specific for the N-terminal region of Zimp10. GAPDH cDNA was amplified as a control. (D) Western blot using affinity-purified anti-Zimp10 antibody to examine the expression of Zimp10 proteins in E9.5 embryos. (E) A schematic representation of the human Zimp10 protein. The numbers correspond to the amino acid sequence of the protein. The putative functional domains are marked as the proline-rich domain (Pro Rich), the nuclear localization signal (NLS), and the Miz domain (Miz).