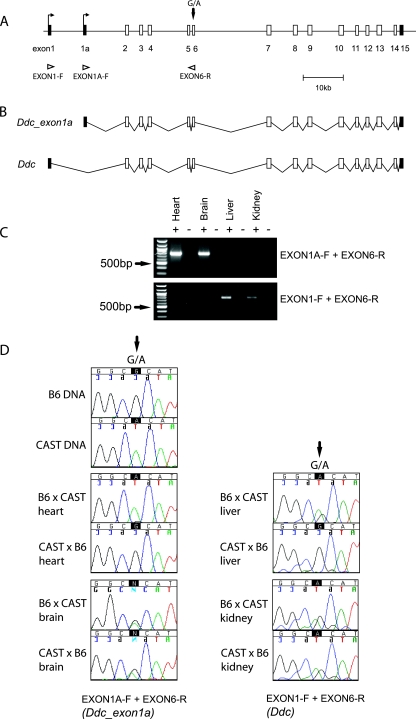
FIG. 2.
Tissue- and transcript-specific imprinting of mouse Ddc. (A) Map of the ∼84-kb genomic region containing the Ddc locus. Coding exons are shown as open boxes, noncoding exons are shown as filled boxes, and open triangles indicate the positions of primers used in allele-specific RT-PCR assays. The location of a G→A SNP in exon 6 between the Mus mus musculus (B6) and Mus mus castaneus (CAST) subspecies is shown by a vertical arrow. (B) Deduced structures of the Ddc_exon1a transcript variant and the related Ddc transcript in their respective genomic contexts. (C) Exon-specific RT-PCR in newborn mouse tissues. The primers EXON1A-F and EXON6-R amplify the Ddc_ exon1a transcript (786-bp product); EXON1-F and EXON6-R amplify the related Ddc transcript (762-bp product). The 500-bp marker is indicated for a reference (arrows). (D) Allele-specific RT-PCR assays in newborn mouse tissues. cDNA fragments were recovered from reciprocal B6 × CAST intersubspecies hybrid tissues by RT-PCR and directly sequenced using the exon-specific primer combinations described above. The G→A SNP (arrows) in exon 6 was used to infer the parental origin of the expressed allele.