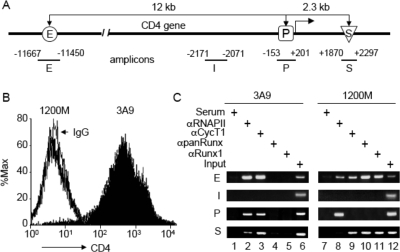
FIG. 1.
Recruitment of RNAPII, P-TEFb (CycT1), and Runx1 to the CD4 locus. (A) Schematic diagram of the CD4 locus and the amplicons used in ChIP assays. The CD4 enhancer (E) is located 12 kb upstream from the promoter (P), and the CD4 silencer (S) is located in the first intron, 2.3 kb downstream from the promoter. I designates the amplicon located between the enhancer and the promoter. (B) FACS analyses of CD4 expression in 1200M (open peak) and 3A9 (solid peak) cells stained with fluorescein isothiocyanate-conjugated anti-mouse CD4 or a rat immunoglobulin G2b(κ) isotype control (thin black line). The intensity of CD4 staining and the relative number of cells are shown on the x and y axes, respectively. (C) ChIP analyses of the CD4 locus in 1200M (CD4−) and 3A9 (CD4+) cells. Anti-RNAPII, anti-CycT1, anti-panRunx, and anti-Runx1 antibodies were used as described in Materials and Methods. Normal rabbit serum served as the negative control for antibody specificity. The presence of the CD4 enhancer, intervening sequence, promoter, and silencer in the immunoprecipitates was examined by PCR with primer sets E, I, P, and S, respectively. PCR analyses with DNA before immunoprecipitation (Input) served as controls for the amplification efficiencies of individual primer sets. PCRs were carried out at various cycle numbers to ensure linear amplification. Representative agarose gels of PCR products within the linear range are presented.