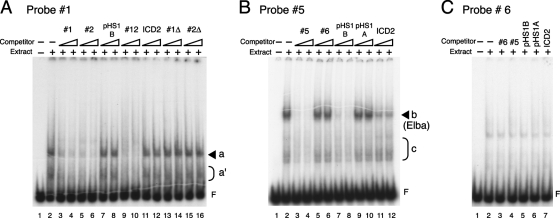
FIG. 2.
Multiple binding activities to the pHS1 region are observed in 0- to 12-h embryo extracts by EMSA. (A) EMSA with labeled probe 1. Probe 1 DNA was 5′ end labeled with 32P and incubated with (lanes 2 to 16) or without (lane 1) 15 μg (protein amount) of nuclear extracts derived from 0- to 12-h embryos in the absence (lanes 1 and 2) or presence (lanes 3 to 16) of unlabeled cold competitor probes as indicated above the lanes. The competitor ICD2 is an 87-bp fragment from Fab-8, used as a heterologous control DNA. Either a 50-fold (left lane of each set) or a 100-fold (right lane of each set) excess of the cold competitor was added to the reaction mixture. After a 30-min incubation at room temperature, the samples were electrophoresed on a 4% acrylamide-0.5× TBE-2.5% glycerol gel. The identity of the shifted band is indicated by a solid arrowhead or half-parentheses on the right. The letter F represents the position of “free”’ (unbound) probe. (B) EMSA with probe 5. End-labeled probe 5 DNA was incubated with (lanes 2 to 12) or without (lane 1) 15 μg (protein amount) of nuclear extracts from 0- to 12-h embryos in the absence (lanes 1 and 2) or presence (lanes 3 to 12) of competitor probes as indicated. Other EMSA experimental conditions were the same as those described for panel A. The free probe and the identities of shifted bands are indicated as described for panel A. (C) EMSA with probe 6. Probe 6 was incubated with (lanes 2 to 7) or without (lane 1) 15 μg (protein amount) of nuclear extracts from 0- to 12-h embryos in the absence (lanes 1 and 2) or presence (lanes 3 to 7) of the competitor probes indicated. A 100-fold excess of unlabeled DNA was used for each competition.