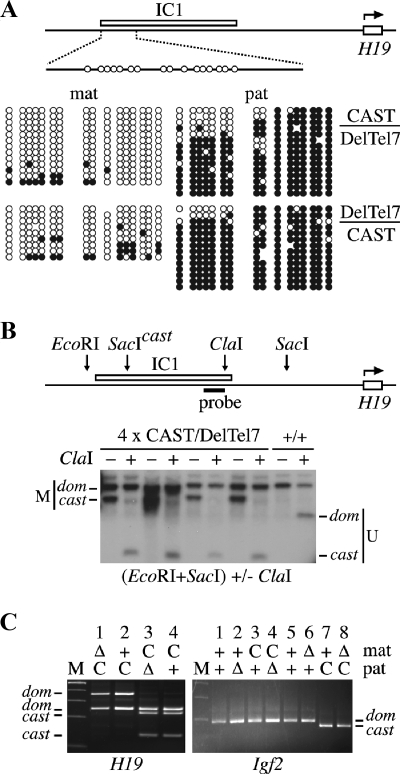
FIG. 6.
Normal imprinting at IC1 in DelTel7 heterozygotes. (A) Diagram of the upstream region of H19, showing the position of the IC1 (H19 DMR) (66) and the structure of a 473-bp sequence containing the 16 CpG sites analyzed by bisulfite sequencing. Individual embryos were recovered at E9.5 from reciprocal crosses between +/DelTel7 and (129CAST)F1 mice. PCR genotyping on yolk sac lysates was used to identify DelTel7 heterozygous embryos carrying the CAST alleles for distal Chr 7. Genomic DNA samples from a CAST/DelTel7 and a DelTel7/CAST embryo were used for bisulfite sequencing analysis of DNA methylation patterns at IC1. Unlike IC2, IC1 and H19 are not deleted in DelTel7 (Fig. 1A). Both parental alleles were analyzed, and SNPs in the sequenced products were used to identify the parental origin of each DNA strand analyzed. Methylated CpG sites are represented by filled circles, and unmethylated sites are shown as open circles. Omitted sites indicate sequencing ambiguities. (B) Southern blot analysis at IC1 for genomic DNA samples from E14.5 CAST/DelTel7 embryos and a wild-type (+/+) littermate. Digestions were with EcoRI and SacI, without (-) or with (+) the methylation-sensitive enzyme ClaI. A CAST SacI polymorphism was used to distinguish maternal and paternal alleles. The diagram shows the positions of the enzyme cut sites relative to IC1 in the H19 5′-flanking region and that of the probe used. Methylated (M) and unmethylated (U) bands are identified for the M. domesticus (dom) and M. castaneus (cast) alleles. (C) Allele-specific expression analysis of H19 and Igf2 in placental RNA samples at E9.5. For each sample, the maternal and paternal alleles are shown as follows: C, wild-type M. castaneus allele; Δ, DelTel7; +, wild-type M. domesticus allele. Note that both Δ and wild-type alleles have M. domesticus variants of H19 and Igf2 on distal Chr 7.