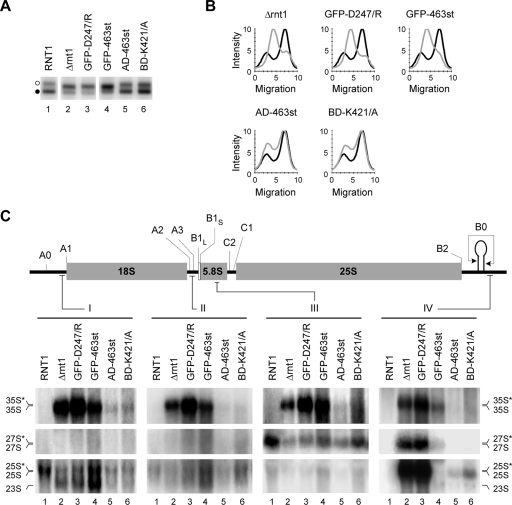
FIG. 6.
Mutations that disrupt the Rnt1p/RNAPI complex increase the number of open rRNA genes and reduce the efficiency of pre-rRNA processing. (A) The ratios of open to closed rRNA genes were examined in cells expressing different mutations that impair Rnt1p catalytic activity (GFP-D247/R), block the localization of Rnt1p in the nucleolus (GFP-463st), or disrupt the interaction with RNAPI (AD-463st and BD-K421/A). The open and closed rRNA genes are indicated by open and black circles, respectively, on the left. The profiles corresponding to the samples shown in panel A are presented in panel B. Wild-type profiles are in black, while those of the different mutants are in gray. (C) Northern blot analyses of rRNAs extracted from cells expressing the different Rnt1p mutants examined in panel A. The pre-rRNA processing intermediates were visualized with probes against sequences in the 5′ ETS, ITS1, 5.8S rRNA, or 3′ ETS (I to IV). Schematic representations of the primary rRNA transcript and probe positions are shown at the top. The positions of the different processing intermediates are indicated at the sides, with asterisks denoting 3′-extended species detected with probe IV.