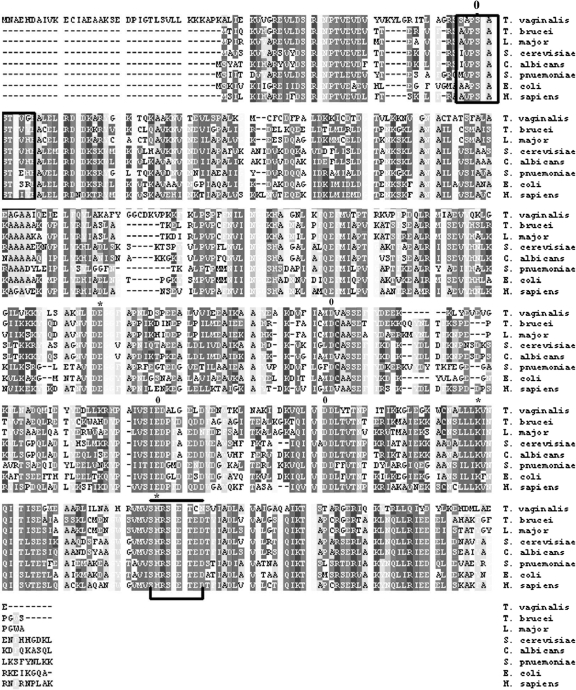
FIG. 1.
Sequence analyses of T. vaginalis α-enolase. T. vaginalis tv-ENO1 was compared to the enolase enzymes of parasites, fungi, bacteria, and humans. The amino acid sequence identity values of T. vaginalis tv-ENO1 to the enolase enzymes of Trypanosoma brucei, Leishmania major, Saccharomyces cerevisiae, Candida albicans, Streptococcus pneumoniae, E. coli, and Homo sapiens as listed vertically were 74%, 69%, 84%, 65%, 76%, 65%, and 67%, respectively. Active sites are marked with an asterisk (amino acids E255, K389, and H417), and the positions of metal-binding residues are labeled zero (amino acids S78, D292, E337, and D364). The enolase finger print motif is boxed within amino acids 75 to 84, and the domain for hydrophobic putative signal peptides is indicated by a bar and a bracket (amino acids 416 to 423). Gaps introduced to maximize alignment are indicated by dashes.