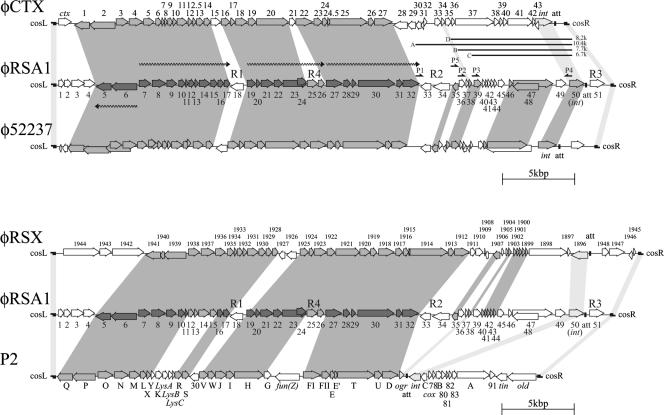
FIG. 1.
(Top) Alignment of φRSA1 ORFs with those of φCTX (38) and φ52237 (accession no. DQ087285). (Bottom) Alignment of φRSA1 ORFs with those of P2 (GenBank accession no. NC001895) and a prophage (designated φRSX in this work) previously found in the genome of R. solanacearum GMI1000 (9, 44). ORFs are indicated by arrows. Predicted transcription units for φRSA1 late genes are shown by wavy lines. Gray shading indicates significant amino acid or nucleotide sequence similarity among the phages. Light gray shading indicates marginally similar regions. R1, R2, R3, and R4 are regions with relatively low G+C contents (AT-rich regions), corresponding to ACURs (44). P1, P2, P3, P5, and P4 are PCR primers for minireplicon formation. A, B, C, and D are fragments of 10.4, 7.7, 6.7, and 8.2 kbp, respectively, used for minireplicon formation.