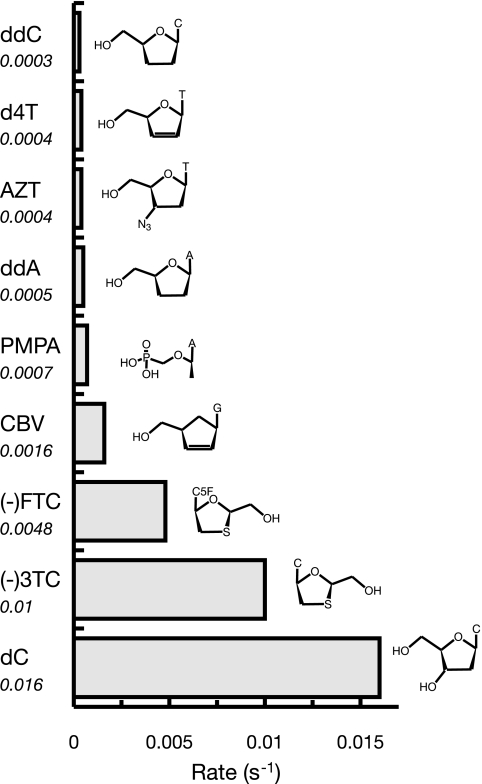
FIG. 4.
Overview of rates of exonuclease removal by Pol γ. Shown in the bar graph are the relative rates of exonuclease removal of the eight FDA-approved nucleoside analogs (ddATP [ddA] is the metabolically active form of dideoxyinosine) in addition to the natural nucleotide dCMP (dC). The rate of excision of ddC was obtained in the experiment with a correctly paired DNA substrate and no dNTPs present so that parallel comparisons could be made. The numerical value for the rate of exonuclease removal (s−1) is plotted on the x axis and given below the name of each nucleoside analog. Note that the chemical structures have been abbreviated for clarity and that the only nucleoside analog with a modified nucleobase is (−)FTC, which has fluorine on C-5, denoted in the structure as C5F. The most notable differences between each of the structures are in the ribose ring, and the highest rate of removal is 33-fold higher than the lowest rate. The rates of removal of d4T, AZT, ddATP, PMPA, and CBV are from reference 15, that of (−)3TC is from reference 7, and that for (−)FTC is from reference 8.