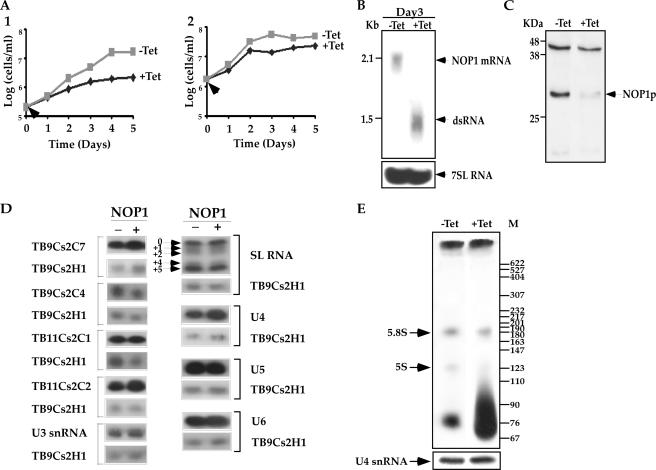
FIG. 1.
NOP1 is essential for life. (A) Growth curves of cells carrying the NOP1 RNAi construct. The growth of uninduced cells (−Tet [gray]) was compared with that of cells in which dsRNA production was induced (+Tet [black]). Arrows indicate the times when tetracycline was added. (Panel 1) Cell growth at 27°C; (panel 2) cell growth at 37°C. (B) Northern blot analysis of NOP1 mRNA upon RNAi silencing. RNA was prepared from cells carrying the RNAi construct, either uninduced (−Tet) or induced for 3 days (+Tet). Total RNA (30 μg) was separated on a 1.2% agarose-2.2 M formaldehyde gel. The RNA was blotted and hybridized with a randomly labeled probe specific for the NOP1 gene (positions 389 to 899 of the coding sequence). The level of 7SL RNA (used as a control for equal loading) was determined using a randomly labeled probe specific for 7SL RNA as described in Fig. S-7 in the supplemental material. Positions of markers are indicated on the left. (C) Western blot analysis of Nop1p. Whole-cell extracts (106 cells per lane) were prepared from cells carrying the RNAi construct, either uninduced (−Tet) or after 3 days of induction (+Tet). The extracts were separated on a 12% SDS-polyacrylamide gel and subjected to Western blot analysis with the anti-fibrillarin antibody (9). (D) Effects of NOP1 silencing on the levels of C/D snoRNAs and snRNAs. Total RNA was prepared from cells carrying the RNAi construct either without induction (−) or after 3 days of induction (+). Total RNA (10 μg) was subjected to primer extension with radiolabeled oligonucleotides complementary to each of the C/D snoRNAs and to SL RNA and snRNAs. The oligonucleotides and their sequences are summarized in Fig. S-7 in the supplemental material. cDNA was separated on a 6% sequencing gel. To control for the level of RNA in each sample, primer extension was performed with primer TBsno-H-1, specific to TB9Cs2H1 H/ACA snoRNA. (E) In vivo labeling of rRNA under NOP1 depletion. Uninduced cells and cells induced for 2 days were labeled with 100 μCi of [methyl-3H]methionine as described in Materials and Methods. RNA was separated on a 6% polyacrylamide-7 M urea gel next to a DNA marker (MspI-digested pBR322). The levels of RNA in the samples were determined by analyzing the U4 snRNA by primer extension.