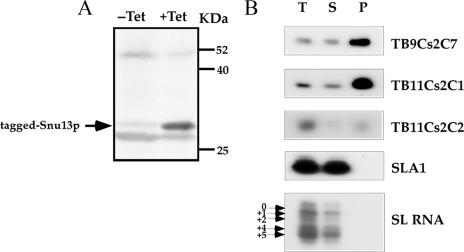
FIG. 9.
Affinity selection of C/D snoRNAs using TAP-tagged Snu13p protein. (A) Western blot analysis. Whole-cell extracts were prepared from uninduced (−Tet) or induced (+Tet) cells (50 ml; 107 cells/ml) carrying the Snu13-TAP construct. The extracts were fractioned on a 12% SDS-polyacrylamide gel and subjected to Western blot analysis with IgG antibodies. Arrow indicates tagged Snu13p. (B) Specific affinity selection of C/D snoRNAs with IgG beads. Whole-cell extracts were prepared from induced cells (500 ml; 107 cells/ml) carrying the Snu13-TAP construct as described in Materials and Methods. The extracts were subjected to affinity selection using IgG-Sepharose beads. RNAs from total-cell extracts (1/30), supernatants (1/30), and all of the selected RNA (lanes T, S, and P, respectively) were subjected to primer extension analysis with radiolabeled oligonucleotides complementary to the designated RNA. The RNA was separated on a 6% denaturing gel. The position of the Cap-4 nt on the SL RNA (the four hypermodified nt at the 5′ end of the SL RNA) is indicated.