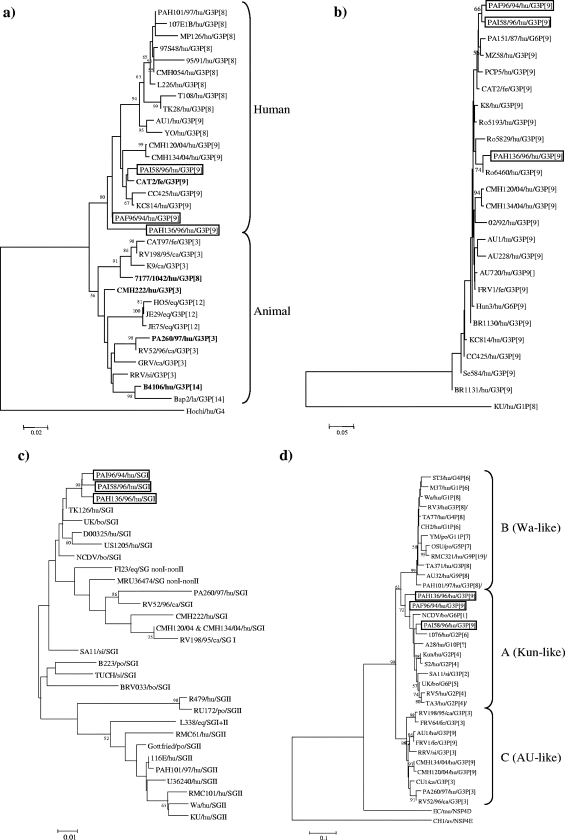
FIG. 1.
Phylogenetic analysis of deduced amino acid sequence of the VP7 (a), VP4 (b), VP6 (c), and NSP4 (d) genes. The phylogeny was inferred by the neighbor-joining method and by use of the Kimura two-parameter model as the method of substitution. The statistical significance was estimated by bootstrap analysis with 1,000 pseudoreplicate data sets. The origins of the rotavirus strains are indicated as follows: hu, human; fe, feline; ca, canine; eq, equine; si, simian; la, lapine; bo, bovine; po, porcine; mu, murine; and av, avian. In panel a, strains with atypical positions in the VP7 tree are highlighted in boldface.