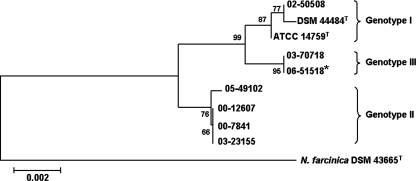
FIG. 2.
Unrooted phylogenetic tree based on the concatenated 16S rRNA and hsp65 gene sequences (1,872 bp in total). Sequences were aligned using Molecular Evolutionary Genetics Analysis software, version 3.1, by ClustalW, and a phylogenetic tree was constructed using the neighbor-joining method and tested for robustness by bootstrapping with 2,000 replicates. N. farcinica strain DSM 43665T was used as the outgroup. The reference bar marks a 0.2% estimated sequence variance. An asterisk denotes the isolate from the index patient.