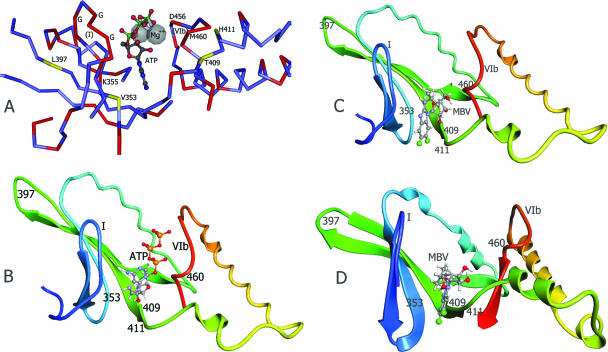
FIG. 3.
Structure models of UL97 kinase ATP and MBV binding sites. (A) Corresponding residues of the UL97 sequence are labeled on the experimentally determined structure of yeast GCN2 kinase complexed to ATP (PDB 1zyd) (24). Residues identical in the two kinases are shown in red on the peptide backbone. G, conserved glycine residues in kinase domain I. (B) UL97 kinase ATP binding site structure model, based on the yeast GCN2 kinase structure (PDB 1zyc) (24) and the sequence alignment shown in Fig. 2A, as constructed by CPH Models 2.0. (C) MBV docked to the same UL97 structure model as above, guided by residues 353, 409, 411, and 460. (D) MBV docked to an alternative UL97 structure model based on the human SRPK1 structure (PDB 1wbp) (23) and the sequence alignment shown in Fig. 2B, as constructed by MODELLER v9.1. In the UL97 structure models, the ATP or MBV molecule was docked using ArgusLab v4 and structure displayed as a ribbon cartoon of protein backbone. Conserved kinase domains I and VIb and residues affecting MBV sensitivity are labeled.